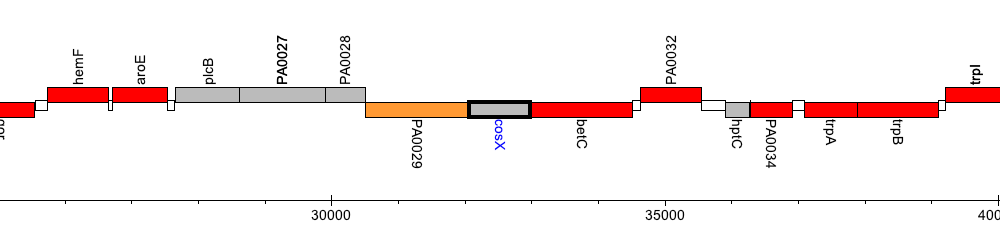
Pseudomonas aeruginosa PAO1, PA0030 (cosX)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0042425 | choline biosynthetic process |
Inferred from Sequence or Structural Similarity
Term mapped from: UniProtKB:Q500S0
|
ECO:0000250 sequence similarity evidence used in manual assertion |
23763788 | Reviewed by curator |
| Cellular Component | GO:0042597 | periplasmic space |
Inferred from Sequence or Structural Similarity
Term mapped from: UniProtKB:Q500S0
|
ECO:0000250 sequence similarity evidence used in manual assertion |
23763788 | Reviewed by curator |
| Cellular Component | GO:0043190 | ATP-binding cassette (ABC) transporter complex |
Inferred from Sequence or Structural Similarity
Term mapped from: UniProtKB:Q500S0
|
ECO:0000250 sequence similarity evidence used in manual assertion |
23763788 | Reviewed by curator |
| Molecular Function | GO:1901682 | sulfur compound transmembrane transporter activity |
Inferred from Sequence or Structural Similarity
Term mapped from: UniProtKB:Q500S0
|
ECO:0000250 sequence similarity evidence used in manual assertion |
23763788 | Reviewed by curator |
| Molecular Function | GO:0033265 | choline binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03414
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0022857 | transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF04069
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0042597 | periplasmic space |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03414
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0043190 | ATP-binding cassette (ABC) transporter complex |
Inferred from Sequence Model
Term mapped from: InterPro:PF04069
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0055085 | transmembrane transport |
Inferred from Sequence Model
Term mapped from: InterPro:PF04069
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0015871 | choline transport |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR03414
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02010 | ABC transporters | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.190.100 | - | - | - | 111 | 220 | 2.2E-92 |
| NCBIfam | TIGR03414 | JCVI: choline ABC transporter substrate-binding protein | IPR017783 | Choline ABC transporter, substrate-binding protein | 22 | 305 | 8.4E-127 |
| Pfam | PF04069 | Substrate binding domain of ABC-type glycine betaine transport system | IPR007210 | ABC-type glycine betaine transport system, substrate-binding domain | 29 | 279 | 1.1E-55 |
| Gene3D | G3DSA:3.40.190.10 | - | - | - | 30 | 288 | 2.2E-92 |
| SUPERFAMILY | SSF53850 | Periplasmic binding protein-like II | - | - | 22 | 295 | 2.35E-81 |
| FunFam | G3DSA:3.40.190.100:FF:000002 | Choline ABC transporter substrate-binding protein | - | - | 111 | 220 | 2.3E-59 |
| CDD | cd13640 | PBP2_ChoX | IPR017783 | Choline ABC transporter, substrate-binding protein | 30 | 291 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.