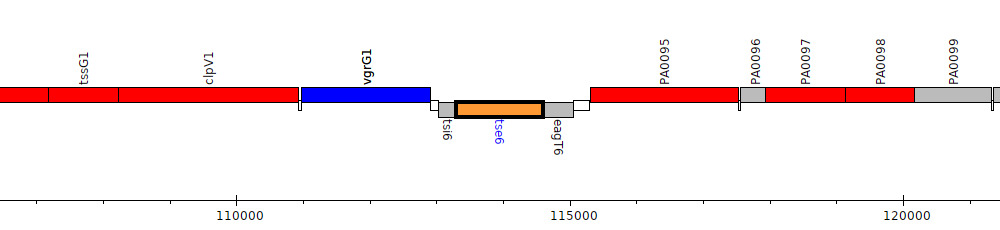
Pseudomonas aeruginosa PAO1, PA0093 (tse6)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009405 | pathogenesis | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
24589350 | Reviewed by curator |
| Biological Process | GO:0033103 | protein secretion by the type VI secretion system | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
24589350 | Reviewed by curator |
| Biological Process | GO:0009405 | pathogenesis | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
24794869 | Reviewed by curator |
| Molecular Function | GO:0050135 | NAD(P)+ nucleosidase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
26456113 | Reviewed by curator |
| Molecular Function | GO:0097351 | toxin-antitoxin pair type II binding | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
24589350 | Reviewed by curator |
| Biological Process | GO:0033103 | protein secretion by the type VI secretion system | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
24794869 | Reviewed by curator |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF15538 | Bacterial toxin 46 | IPR028238 | Bacterial toxin 46 | 266 | 422 | 3.2E-97 |
| Gene3D | G3DSA:2.60.200.60 | - | - | - | 70 | 161 | 1.2E-20 |
| CDD | cd14742 | PAAR_RHS | - | - | 80 | 161 | 1.4646E-28 |
| Pfam | PF05488 | PAAR motif | IPR008727 | PAAR motif | 80 | 151 | 3.9E-12 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.