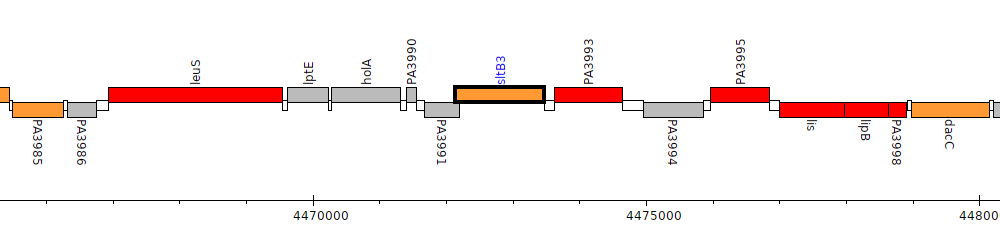
Pseudomonas aeruginosa PAO1, PA3992 (sltB3)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0008933 | lytic transglycosylase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
27035839 | Reviewed by curator |
| Biological Process | GO:0009254 | peptidoglycan turnover | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
27035839 | Reviewed by curator |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF13406 | Transglycosylase SLT domain | IPR031304 | Transglycosylase SLT domain 2 | 75 | 369 | 1.3E-110 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 49 | 74 | - |
| PANTHER | PTHR30163 | MEMBRANE-BOUND LYTIC MUREIN TRANSGLYCOSYLASE B | IPR043426 | Membrane-bound lytic murein transglycosylase B-like | 27 | 439 | 0.0 |
| FunFam | G3DSA:1.10.8.350:FF:000001 | Lytic murein transglycosylase B | - | - | 171 | 251 | 4.8E-26 |
| SUPERFAMILY | SSF53955 | Lysozyme-like | IPR023346 | Lysozyme-like domain superfamily | 68 | 375 | 3.75E-104 |
| SUPERFAMILY | SSF47090 | PGBD-like | IPR036365 | PGBD-like superfamily | 377 | 446 | 3.4E-17 |
| Gene3D | G3DSA:1.10.530.10 | - | - | - | 118 | 368 | 2.0E-83 |
| CDD | cd13399 | Slt35-like | - | - | 158 | 287 | 2.16796E-24 |
| Gene3D | G3DSA:1.10.101.10 | - | IPR036366 | PGBD superfamily | 389 | 447 | 6.3E-12 |
| NCBIfam | TIGR02283 | JCVI: lytic murein transglycosylase | IPR011970 | Lytic murein transglycosylase | 75 | 373 | 2.2E-117 |
| Pfam | PF01471 | Putative peptidoglycan binding domain | IPR002477 | Peptidoglycan binding-like | 391 | 445 | 1.7E-12 |
| Gene3D | G3DSA:1.10.8.350 | Bacterial muramidase | - | - | 91 | 251 | 2.0E-83 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.