Pseudomonas aeruginosa PAO1, PA4921 (choE)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
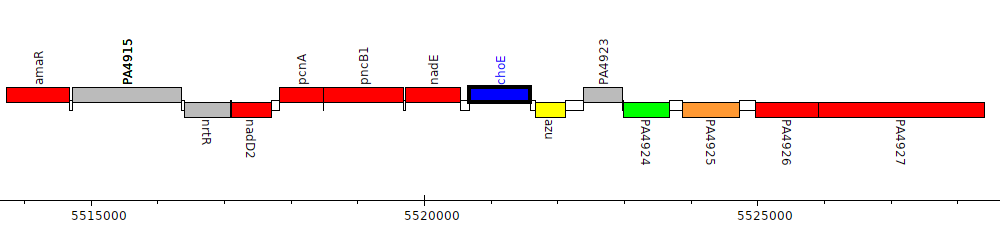
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4921
|
| Name |
choE
|
| Replicon | chromosome |
| Genomic location | 5520672 - 5521595 (+ strand) |
| Transposon Mutants | 1 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 3 transposon mutants in orthologs |
| Comment | Gene changed names between the authors' original submission and the paper acceptance. |
Cross-References
| RefSeq | NP_253608.1 |
| GI | 15600114 |
| Affymetrix | PA4921_at |
| Entrez | 878045 |
| GenBank | AAG08306.1 |
| INSDC | AHL26984.1 |
| NCBI Locus Tag | PA4921 |
| protein_id(GenBank) | gb|AAG08306.1|AE004905_4|gnl|PseudoCAP|PA4921 |
| TIGR | NTL03PA04922 |
| UniParc | UPI00000C5E7D |
| UniProtKB Acc | Q9HUP2 |
| UniProtKB ID | Q9HUP2_PSEAE |
| UniRef100 | UniRef100_Q9HUP2 |
| UniRef50 | UniRef50_Q9HUP2 |
| UniRef90 | UniRef90_Q9HUP2 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
cholinesterase, ChoE
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | 4.31 |
| Kyte-Doolittle Hydrophobicity Value | -0.227 |
| Molecular Weight (kDa) | 33.5 |
| Isoelectric Point (pI) | 8.46 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 6UQY | HYDROLASE | 10/21/19 | Crystal structure of ChoE H288N mutant in complex with acetylthiocholine | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.57 | X-RAY DIFFRACTION | 99.7 |
| 6UQX | HYDROLASE | 10/21/19 | Crystal structure of ChoE in complex with propionylthiocholine | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.85 | X-RAY DIFFRACTION | 100.0 |
| 6UQV | HYDROLASE | 10/21/19 | Crystal structure of ChoE, a bacterial acetylcholinesterase from Pseudomonas aeruginosa | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.35 | X-RAY DIFFRACTION | 100.0 |
| 8D8W | HYDROLASE | 06/09/22 | Crystal structure of ChoE with Ser38 adopting alternative conformations | Pseudomonas aeruginosa | 1.4 | X-RAY DIFFRACTION | 100.0 |
| 6UR0 | HYDROLASE | 10/21/19 | Crystal structure of ChoE D285N mutant acyl-enzyme | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.8 | X-RAY DIFFRACTION | 99.3 |
| 8D8Y | HYDROLASE | 06/09/22 | Crystal structure of ChoE N147A mutant in complex with acetylthiocholine | Pseudomonas aeruginosa | 1.54 | X-RAY DIFFRACTION | 99.7 |
| 6UQW | HYDROLASE | 10/21/19 | Crystal structure of ChoE in complex with acetate and thiocholine | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.65 | X-RAY DIFFRACTION | 100.0 |
| 8D8Z | HYDROLASE | 06/09/22 | Crystal structure of ChoE N147A mutant in complex with thiocholine and chloride | Pseudomonas aeruginosa | 1.38 | X-RAY DIFFRACTION | 99.7 |
| 8D90 | HYDROLASE | 06/09/22 | Crystal structure of ChoE N147A mutant in complex with bromide ions | Pseudomonas aeruginosa | 1.82 | X-RAY DIFFRACTION | 99.7 |
| 6UQZ | HYDROLASE | 10/21/19 | Crystal structure of ChoE D285N mutant in complex with acetate and thiocholine | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.43 | X-RAY DIFFRACTION | 99.7 |
| 6UR1 | HYDROLASE | 10/21/19 | Crystal structure of ChoE S38A mutant in complex with acetate and acetylthiocholine | Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) | 1.42 | X-RAY DIFFRACTION | 99.7 |
| 8D8X | HYDROLASE | 06/09/22 | Crystal structure of ChoE in complex with acetate and thiocholine (crystal form 2) | Pseudomonas aeruginosa | 1.36 | X-RAY DIFFRACTION | 100.0 |
| 8D91 | HYDROLASE | 06/09/22 | Crystal structure of ChoE in complex with acetate and tetraethylammonium (TEA) | Pseudomonas aeruginosa | 1.49 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 72 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG004538 (814 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4921
Search term: choE
Search term: cholinesterase, ChoE
|
Human Homologs
References
|
Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen.
Lewenza S, Gardy JL, Brinkman FS, Hancock RE
Genome Res 2005 Feb;15(2):321-9
PubMed ID: 15687295
|
|
Molecular cloning of a phospholipid-cholesterol acyltransferase from Aeromonas hydrophila. Sequence homologies with lecithin-cholesterol acyltransferase and other lipases.
Thornton J, Howard SP, Buckley JT
Biochim Biophys Acta 1988 Mar 25;959(2):153-9
PubMed ID: 3280033
|
|
A Pseudomonas aeruginosa PAO1 acetylcholinesterase is encoded by the PA4921 gene and belongs to the SGNH hydrolase family.
Sánchez DG, Otero LH, Hernández CM, Serra AL, Encarnación S, Domenech CE, Lisa AT
Microbiol Res 2012 Jun 20;167(6):317-25
PubMed ID: 22192836
|