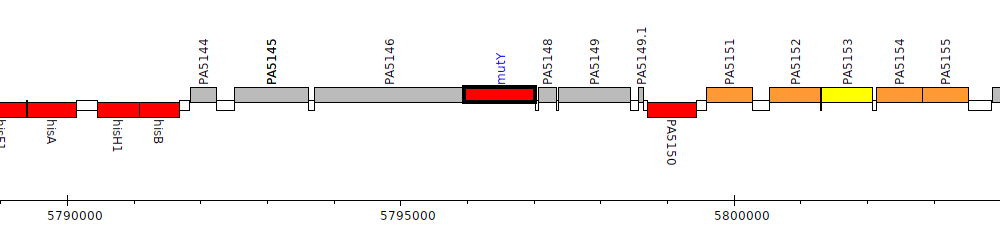
Pseudomonas aeruginosa PAO1, PA5147 (mutY)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA5147
|
| Name |
mutY
Synonym: mutM Synonym: micA |
| Replicon | chromosome |
| Genomic location | 5795954 - 5797021 (+ strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_253834.1 |
| GI | 15600340 |
| Affymetrix | PA5147_mutY_at |
| Entrez | 878753 |
| GenBank | AAG08532.1 |
| INSDC | AAG08532.1 |
| NCBI Locus Tag | PA5147 |
| protein_id(GenBank) | gb|AAG08532.1|AE004927_10|gnl|PseudoCAP|PA5147 |
| TIGR | NTL03PA05148 |
| UniParc | UPI00000C5F25 |
| UniProtKB Acc | Q9HU37 |
| UniProtKB ID | Q9HU37_PSEAE |
| UniRef100 | UniRef100_Q9HU37 |
| UniRef50 | UniRef50_Q9HU37 |
| UniRef90 | UniRef90_Q9HU37 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
A / G specific adenine glycosylase
|
| Synonyms | |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | 5.29 |
| Kyte-Doolittle Hydrophobicity Value | -0.213 |
| Molecular Weight (kDa) | 39.3 |
| Isoelectric Point (pI) | 8.59 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 621 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG004741 (533 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA5147
Search term: mutY
Search term: A / G specific adenine glycosylase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
novel protein
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
|
Ensembl
110, assembly
GRCh38.p14
|
mutY DNA glycosylase [Source:HGNC Symbol;Acc:HGNC:7527]
E-value:
1.1e-53
Percent Identity:
36.7
|
References
|
MutY, an adenine glycosylase active on G-A mispairs, has homology to endonuclease III.
Michaels ML, Pham L, Nghiem Y, Cruz C, Miller JH
Nucleic Acids Res 1990 Jul 11;18(13):3841-5
PubMed ID: 2197596
|
|
Nucleotide sequence of the Escherichia coli micA gene required for A/G-specific mismatch repair: identity of micA and mutY.
Tsai-Wu JJ, Radicella JP, Lu AL
J Bacteriol 1991 Mar;173(6):1902-10
PubMed ID: 2001994
|
|
Characterization of the GO system of Pseudomonas aeruginosa.
Oliver A, Sánchez JM, Blázquez J
FEMS Microbiol Lett 2002 Nov 19;217(1):31-5
PubMed ID: 12445642
|