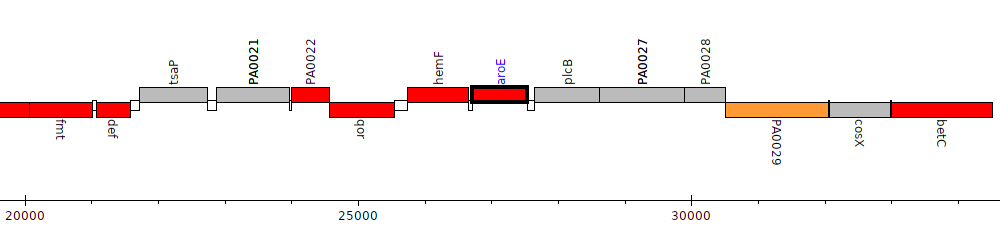
Pseudomonas aeruginosa PAO1, PA0025 (aroE)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006571 | tyrosine biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
3277621 | Reviewed by curator |
| Biological Process | GO:0009094 | L-phenylalanine biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
3277621 | Reviewed by curator |
| Molecular Function | GO:0004764 | shikimate 3-dehydrogenase (NADP+) activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
3277621 | Reviewed by curator |
| Biological Process | GO:0006520 | cellular amino acid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
3277621 | Reviewed by curator |
| Biological Process | GO:0000162 | tryptophan biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
3277621 | Reviewed by curator |
| Molecular Function | GO:0050661 | NADP binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00507
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0019632 | shikimate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00507
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004764 | shikimate 3-dehydrogenase (NADP+) activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00507
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCyc | ALL-CHORISMATE-PWY | superpathway of chorismate metabolism | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01230 | Biosynthesis of amino acids | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | ARO-PWY | chorismate biosynthesis I | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY-6163 | chorismate biosynthesis from 3-dehydroquinate | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCAP | Phenylalanine, tyrosine and tryptophan biosynthesis |
ECO:0000037
not_recorded |
|||
| PseudoCyc | COMPLETE-ARO-PWY | superpathway of aromatic amino acid biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Hamap | MF_00222 | Shikimate dehydrogenase (NADP(+)) [aroE]. | IPR022893 | Shikimate dehydrogenase family | 1 | 270 | 31.42564 |
| FunFam | G3DSA:3.40.50.720:FF:000104 | Shikimate dehydrogenase (NADP(+)) | - | - | 101 | 245 | 6.8E-60 |
| SUPERFAMILY | SSF53223 | Aminoacid dehydrogenase-like, N-terminal domain | IPR046346 | Aminoacid dehydrogenase-like, N-terminal domain superfamily | 1 | 100 | 1.09E-30 |
| NCBIfam | TIGR00507 | JCVI: shikimate dehydrogenase | IPR011342 | Shikimate dehydrogenase | 4 | 271 | 9.4E-76 |
| SUPERFAMILY | SSF51735 | NAD(P)-binding Rossmann-fold domains | IPR036291 | NAD(P)-binding domain superfamily | 101 | 270 | 1.25E-36 |
| Pfam | PF01488 | Shikimate / quinate 5-dehydrogenase | IPR006151 | Quinate/shikimate 5-dehydrogenase/glutamyl-tRNA reductase | 116 | 165 | 4.7E-7 |
| CDD | cd01065 | NAD_bind_Shikimate_DH | - | - | 100 | 255 | 2.5394E-40 |
| Pfam | PF18317 | Shikimate 5'-dehydrogenase C-terminal domain | IPR041121 | SDH, C-terminal | 238 | 267 | 3.4E-11 |
| FunFam | G3DSA:3.40.50.10860:FF:000006 | Shikimate dehydrogenase (NADP(+)) | - | - | 5 | 120 | 2.1E-42 |
| Gene3D | G3DSA:3.40.50.10860 | Leucine Dehydrogenase, chain A, domain 1 | - | - | 6 | 255 | 6.0E-87 |
| Pfam | PF08501 | Shikimate dehydrogenase substrate binding domain | IPR013708 | Shikimate dehydrogenase substrate binding, N-terminal | 6 | 87 | 5.5E-24 |
| Gene3D | G3DSA:3.40.50.720 | - | - | - | 101 | 245 | 6.0E-87 |
| PANTHER | PTHR21089 | SHIKIMATE DEHYDROGENASE | IPR022893 | Shikimate dehydrogenase family | 2 | 259 | 5.0E-60 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.