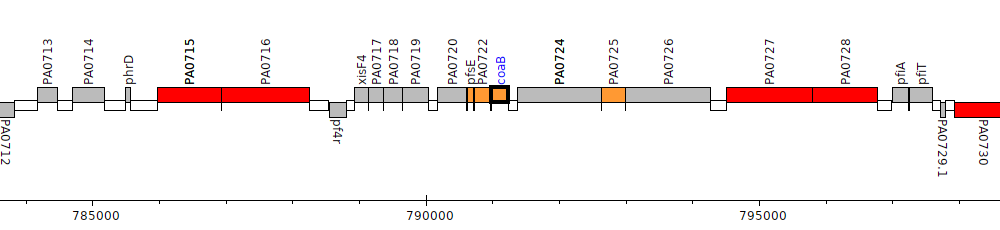
Pseudomonas aeruginosa PAO1, PA0723 (coaB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0723
|
| Name |
coaB
|
| Replicon | chromosome |
| Genomic location | 790986 - 791234 (+ strand) |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_249414.1 |
| GI | 15595920 |
| Affymetrix | PA0723_coaB_at |
| DNASU | PaCD00006332 |
| Entrez | 880778 |
| GenBank | AAG04112.1 |
| INSDC | ACS36651.1 |
| NCBI Locus Tag | PA0723 |
| protein_id(GenBank) | gb|AAG04112.1|AE004507_11|gnl|PseudoCAP|PA0723 |
| TIGR | NTL03PA00724 |
| UniParc | UPI00000C511F |
| UniProtKB Acc | Q9I5K5 |
| UniProtKB Acc | Q9I5K5 |
| UniProtKB ID | C6JW45_PSEAE |
| UniProtKB ID | Q9I5K5_PSEAE |
| UniRef100 | UniRef100_P03621 |
| UniRef50 | UniRef50_P03621 |
| UniRef90 | UniRef90_P03621 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
coat protein B of bacteriophage Pf1
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | 3.95 |
| Kyte-Doolittle Hydrophobicity Value | 0.717 |
| Molecular Weight (kDa) | 8.4 |
| Isoelectric Point (pI) | 10.17 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
AlphaFold 2 Protein Structure Predictions
Protein structure predictions using a neural network model developed by DeepMind. If a UniProtKB accession is associated with this protein, a search link will be provided below.
| Look for predicted 3D structure in AlphaFold DB: Search |
| Look for predicted 3D structure in AlphaFold DB: Search |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 1IFM | VIRUS | 01/31/94 | TWO FORMS OF PF1 INOVIRUS: X-RAY DIFFRACTION STUDIES ON A STRUCTURAL PHASE TRANSITION AND A CALCULATED LIBRATION NORMAL MODE OF THE ASYMMETRIC UNIT | Pseudomonas phage Pf1 | 3.3 | FIBER DIFFRACTION | 100.0 |
| 1ZN5 | VIRUS | 05/11/05 | Solid State NMR Structure of the low-temperature form of the Pf1 Major Coat Protein in Magnetically Aligned Bacteriophage | Pseudomonas phage Pf1 | NOT | SOLID-STATE NMR | 100.0 |
| 1PFI | VIRUS | 04/06/94 | PF1 VIRUS STRUCTURE: HELICAL COAT PROTEIN AND DNA WITH PARAXIAL PHOSPHATES | Pseudomonas phage Pf1 | 3 | FIBER DIFFRACTION | 100.0 |
| 6TUQ | VIRUS | 01/08/20 | Cryo-EM structure of Pf4 bacteriophage coat protein without ssDNA | Pseudomonas virus Pf1 | 3.9 | ELECTRON MICROSCOPY | 100.0 |
| 6TUP | VIRUS | 01/08/20 | Cryo-EM structure of Pf4 bacteriophage coat protein with single-stranded DNA | Pseudomonas aeruginosa PAO1; Pseudomonas virus Pf1 | 3.2 | ELECTRON MICROSCOPY | 100.0 |
| 2KLV | MEMBRANE PROTEIN | 07/08/09 | Membrane-bound structure of the Pf1 major coat protein in DHPC micelle | Pseudomonas phage Pf1 | NOT | SOLUTION NMR | 100.0 |
| 1QL2 | VIRUS | 08/20/99 | Inovirus (Filamentous Bacteriophage) Strain PF1 Major Coat Protein Assembly | PSEUDOMONAS PHAGE PF1 | 3.1 | FIBER DIFFRACTION | 100.0 |
| 1PJF | VIRAL PROTEIN | 06/02/03 | Solid State NMR structure of the Pf1 Major Coat Protein in Magnetically Aligned Bacteriophage | Pseudomonas phage Pf1 | NOT | SOLID-STATE NMR | 100.0 |
| 4IFM | VIRUS | 01/16/95 | PF1 FILAMENTOUS BACTERIOPHAGE: REFINEMENT OF A MOLECULAR MODEL BY SIMULATED ANNEALING USING 3.3 ANGSTROMS RESOLUTION X-RAY FIBRE DIFFRACTION DATA | Pseudomonas phage Pf1 | 3.3 | FIBER DIFFRACTION | 100.0 |
| 2IFN | VIRUS | 01/16/94 | PF1 FILAMENTOUS BACTERIOPHAGE: REFINEMENT OF A MOLECULAR MODEL BY SIMULATED ANNEALING USING 3.3 ANGSTROMS RESOLUTION X-RAY FIBRE DIFFRACTION DATA | Pseudomonas phage Pf1 | 4 | FIBER DIFFRACTION | 100.0 |
| 2IFM | VIRUS | 01/16/95 | PF1 FILAMENTOUS BACTERIOPHAGE: REFINEMENT OF A MOLECULAR MODEL BY SIMULATED ANNEALING USING 3.3 ANGSTROMS RESOLUTION X-RAY FIBRE DIFFRACTION DATA | Xanthomonas phage Xf | 3.3 | FIBER DIFFRACTION | 100.0 |
| 1QL1 | VIRUS | 08/20/99 | INOVIRUS (FILAMENTOUS BACTERIOPHAGE) STRAIN PF1 MAJOR COAT PROTEIN ASSEMBLY | PSEUDOMONAS PHAGE PF1 | 3.1 | FIBER DIFFRACTION | 100.0 |
| 3IFM | VIRUS | 01/16/94 | PF1 FILAMENTOUS BACTERIOPHAGE: REFINEMENT OF A MOLECULAR MODEL BY SIMULATED ANNEALING USING 3.3 ANGSTROMS RESOLUTION X-RAY FIBRE DIFFRACTION DATA | Pseudomonas phage Pf1 | 3.3 | FIBER DIFFRACTION | 100.0 |
| 2XKM | VIRAL PROTEIN | 07/09/10 | Consensus structure of Pf1 filamentous bacteriophage from X-ray fibre diffraction and solid-state NMR | PSEUDOMONAS PHAGE PF1 | 3.3 | FIBER DIFFRACTION, SOLID-STATE NMR | 100.0 |
| 2KSJ | VIRAL PROTEIN | 01/05/10 | Structure and Dynamics of the Membrane-bound form of Pf1 Coat Protein: Implications for Structural Rearrangement During Virus Assembly | Pseudomonas phage Pf1 | NOT | SOLUTION NMR, SOLID-STATE NMR | 100.0 |
| 1IFN | VIRUS | 01/31/94 | TWO FORMS OF PF1 INOVIRUS: X-RAY DIFFRACTION STUDIES ON A STRUCTURAL PHASE TRANSITION AND A CALCULATED LIBRATION NORMAL MODE OF THE ASYMMETRIC UNIT | Pseudomonas phage Pf1 | 4 | FIBER DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in one genus
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000699 (125 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0723
Search term: coaB
Search term: coat protein B of bacteriophage Pf1
|
Human Homologs
References
|
Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen.
Lewenza S, Gardy JL, Brinkman FS, Hancock RE
Genome Res 2005 Feb;15(2):321-9
PubMed ID: 15687295
|
|
Primary structure and sidechain interactions of PFL filamentous bacterial virus coat protein.
Nakashima Y, Wiseman RL, Konigsberg W, Marvin DA
Nature 1975 Jan 3;253(5486):68-71
PubMed ID: 1110754
|