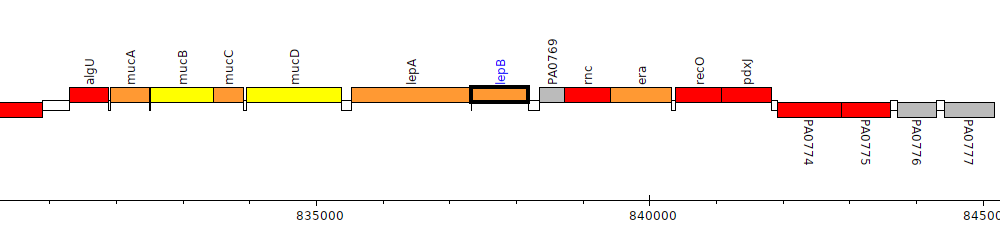
Pseudomonas aeruginosa PAO1, PA0768 (lepB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0768
|
| Name |
lepB
Synonym: lep Synonym: SPASE I |
| Replicon | chromosome |
| Genomic location | 837328 - 838182 (+ strand) |
| Transposon Mutants | 1 transposon mutants in PAO1 |
Cross-References
| RefSeq | NP_249459.1 |
| GI | 15595965 |
| Affymetrix | PA0768_lepB_at |
| Entrez | 882104 |
| GenBank | AAG04157.1 |
| INSDC | AAG04157.1 |
| NCBI Locus Tag | PA0768 |
| protein_id(GenBank) | gb|AAG04157.1|AE004511_10|gnl|PseudoCAP|PA0768 |
| TIGR | NTL03PA00769 |
| UniParc | UPI000012E4A6 |
| UniProtKB Acc | Q9I5G7 |
| UniProtKB ID | LEP_PSEAE |
| UniRef100 | UniRef100_Q9I5G7 |
| UniRef50 | UniRef50_Q9I5G7 |
| UniRef90 | UniRef90_Q9I5G7 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
signal peptidase I
|
| Synonyms |
leader peptidase 1 |
| Evidence for Translation |
Detected using LC-MS/MS (32.10 kDa, PMID:21751344)
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | 2.69 |
| Kyte-Doolittle Hydrophobicity Value | -0.067 |
| Molecular Weight (kDa) | 32.1 |
| Isoelectric Point (pI) | 8.82 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 569 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000742 (535 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0768
Search term: lepB
Search term: signal peptidase I
|
Human Homologs
References
|
Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen.
Lewenza S, Gardy JL, Brinkman FS, Hancock RE
Genome Res 2005 Feb;15(2):321-9
PubMed ID: 15687295
|
|
Sequence of the leader peptidase gene of Escherichia coli and the orientation of leader peptidase in the bacterial envelope.
Wolfe PB, Wickner W, Goodman JM
J Biol Chem 1983 Oct 10;258(19):12073-80
PubMed ID: 6311837
|
|
On the catalytic mechanism of prokaryotic leader peptidase 1.
Black MT, Munn JG, Allsop AE
Biochem J 1992 Mar 1;282 ( Pt 2)(Pt 2):539-43
PubMed ID: 1546969
|
|
Proteomic analysis of outer membrane vesicles derived from Pseudomonas aeruginosa.
Choi DS, Kim DK, Choi SJ, Lee J, Choi JP, Rho S, Park SH, Kim YK, Hwang D, Gho YS
Proteomics 2011 Aug;11(16):3424-9
PubMed ID: 21751344
|
|
Pseudomonas aeruginosa possesses two putative type I signal peptidases, LepB and PA1303, each with distinct roles in physiology and virulence.
Waite RD, Rose RS, Rangarajan M, Aduse-Opoku J, Hashim A, Curtis MA
J Bacteriol 2012 Sep;194(17):4521-36
PubMed ID: 22730125
|