Pseudomonas aeruginosa PAO1, PA1818 (cadA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA1818
|
| Name |
cadA
Synonym: ldcA Synonym: adi Synonym: ldcC Synonym: speC |
| Replicon | chromosome |
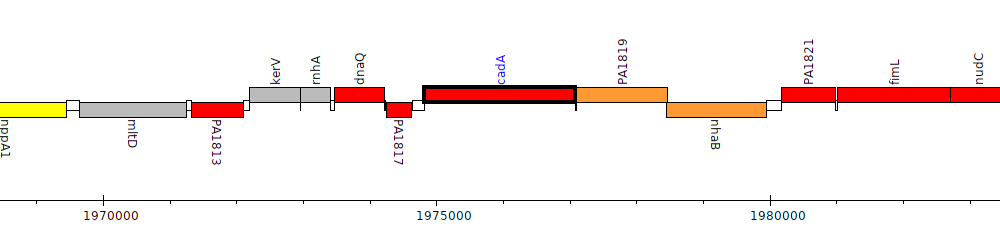
| Genomic location | 1974821 - 1977076 (+ strand) |
| Transposon Mutants | 3 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
| Comment | Modeling evidence, since PAO1 grows on lysine. Also physiological evidence from Fothergill et al. BLAST of Lysine decarboxylase Burkholderia cenocepacia HI2424 vs. P. aeruginosa PAO1 genome gave E = 0 with PA1818. Orn and Arg functions are also in reconstruction. |
Cross-References
| RefSeq | NP_250509.1 |
| GI | 15597015 |
| Affymetrix | PA1818_at |
| Entrez | 878596 |
| GenBank | AAG05207.1 |
| INSDC | AAG05207.1 |
| NCBI Locus Tag | PA1818 |
| protein_id(GenBank) | gb|AAG05207.1|AE004608_6|gnl|PseudoCAP|PA1818 |
| TIGR | NTL03PA01819 |
| UniParc | UPI00000C54AF |
| UniProtKB Acc | Q9I2S7 |
| UniProtKB ID | Q9I2S7_PSEAE |
| UniRef100 | UniRef100_Q9I2S7 |
| UniRef50 | UniRef50_I7KDU0 |
| UniRef90 | UniRef90_Q9I2S7 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
lysine decarboxylase
|
| Synonyms |
Orn/Arg/Lys decarboxylase LdcC lysine-specific pyridoxal 5'-phosphate-dependent carboxylase, LdcA lysine-specific pyridoxal 5’-phosphate-dependent carboxylase, LdcA |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -13.13 |
| Kyte-Doolittle Hydrophobicity Value | -0.083 |
| Molecular Weight (kDa) | 82.8 |
| Isoelectric Point (pI) | 6.07 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 6Q6I | OXIDOREDUCTASE | 12/11/18 | Lysine decarboxylase A from Pseudomonas aeruginosa | Pseudomonas aeruginosa | 3.7 | ELECTRON MICROSCOPY | 99.9 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 242 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG003580 (469 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA1818
Search term: cadA
Search term: lysine decarboxylase
|
Human Homologs
References
|
Catabolism of L-lysine by Pseudomonas aeruginosa.
Fothergill JC, Guest JR
J Gen Microbiol 1977 Mar;99(1):139-55
PubMed ID: 405455
|
|
CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa.
Perron K, Caille O, Rossier C, Van Delden C, Dumas JL, Köhler T
J Biol Chem 2004 Mar 5;279(10):8761-8
PubMed ID: 14679195
|
|
Genes and enzymes of lysine catabolism in Pseudomonas aeruginosa.
Rahman M, Clarke PH
J Gen Microbiol 1980 Feb;116(2):357-69
PubMed ID: 6768834
|
|
L-lysine catabolism is controlled by L-arginine and ArgR in Pseudomonas aeruginosa PAO1.
Chou HT, Hegazy M, Lu CD
J Bacteriol 2010 Nov;192(22):5874-80
PubMed ID: 20833801
|
|
Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440.
Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL
J Bacteriol 2005 Nov;187(21):7500-10
PubMed ID: 16237033
|