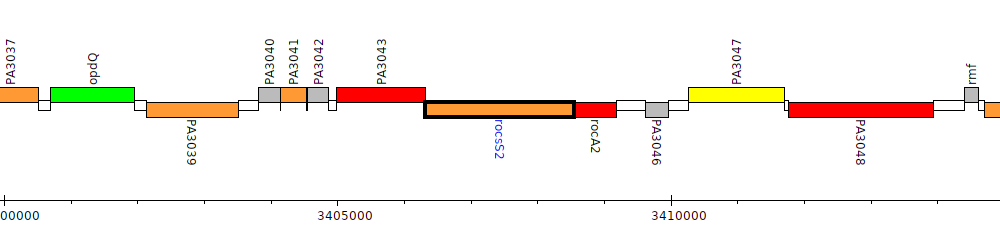
Pseudomonas aeruginosa PAO1, PA3044 (rocsS2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3044
|
| Name |
rocsS2
|
| Replicon | chromosome |
| Genomic location | 3406321 - 3408546 (- strand) |
| Transposon Mutants | 5 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
| Comment | Overexpression of rocS2 leads to the production of CupB and CupC fimbriae, as well as repression of the MexAB-OprM efflux pump. RocS2 can interact with three response regulators: RocA2 (PA3045), RocA1 (PA3948) and RocR (PA3947). |
| Comment | Functional studies in Pseudomonas aeruginosa PAK have shown that RocS2 is part of a signalling network that induces the expression of the cupB and cupC gene clusters, leading to the production and assembly of two distinct types of fimbriae on the cell surface and increases biofilm formation. Expression of rocS2 also represses the expression of the mexAB-oprM gene cluster, leading to increased sensitivity to beta-lactams and other antibiotics. This signalling network involves at least three response regulators: RocA1 (PA3948), RocA2 (PA3045) and RocR (PA3947). Genetic and bacterial two hybrid analyses have shown that RocS2 signals through all three response regulators. |
Cross-References
| RefSeq | NP_251734.1 |
| GI | 15598240 |
| Affymetrix | PA3044_at |
| Entrez | 882873 |
| GenBank | AAG06432.1 |
| INSDC | AAG06432.1 |
| NCBI Locus Tag | PA3044 |
| protein_id(GenBank) | gb|AAG06432.1|AE004729_6|gnl|PseudoCAP|PA3044 |
| TIGR | NTL03PA03044 |
| UniParc | UPI00000C58E4 |
| UniProtKB Acc | Q9HZG4 |
| UniProtKB ID | Q9HZG4_PSEAE |
| UniRef100 | UniRef100_Q9HZG4 |
| UniRef50 | UniRef50_A6V332 |
| UniRef90 | UniRef90_Q9HZG4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
Two-component sensor RocS2
|
| Synonyms | |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | 2.22 |
| Kyte-Doolittle Hydrophobicity Value | -0.199 |
| Molecular Weight (kDa) | 82.2 |
| Isoelectric Point (pI) | 7.40 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 620 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002543 (301 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3044
Search term: rocsS2
Search term: Two-component sensor RocS2
|
Human Homologs
References
|
Genome-wide identification of Pseudomonas aeruginosa exported proteins using a consensus computational strategy combined with a laboratory-based PhoA fusion screen.
Lewenza S, Gardy JL, Brinkman FS, Hancock RE
Genome Res 2005 Feb;15(2):321-9
PubMed ID: 15687295
|
|
A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes.
Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, Lory S
Mol Microbiol 2005 Jan;55(2):368-80
PubMed ID: 15659157
|
|
Two-component regulatory systems in Pseudomonas aeruginosa: an intricate network mediating fimbrial and efflux pump gene expression.
Sivaneson M, Mikkelsen H, Ventre I, Bordi C, Filloux A
Mol Microbiol 2011 Mar;79(5):1353-66
PubMed ID: 21205015
|