Pseudomonas aeruginosa PAO1, PA3484 (tse3)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
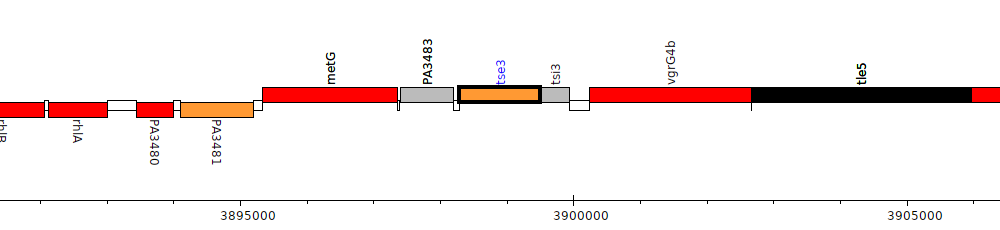
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3484
|
| Name |
tse3
|
| Replicon | chromosome |
| Genomic location | 3898278 - 3899504 (+ strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252174.1 |
| GI | 15598680 |
| Affymetrix | PA3484_at |
| Entrez | 880027 |
| GenBank | AAG06872.1 |
| INSDC | AAG06872.1 |
| NCBI Locus Tag | PA3484 |
| protein_id(GenBank) | gb|AAG06872.1|AE004769_5|gnl|PseudoCAP|PA3484 |
| TIGR | NTL03PA03484 |
| UniParc | UPI00000C5A32 |
| UniProtKB Acc | Q9HYC5 |
| UniProtKB ID | Q9HYC5_PSEAE |
| UniRef100 | UniRef100_Q9HYC5 |
| UniRef50 | UniRef50_Q9HYC5 |
| UniRef90 | UniRef90_Q9HYC5 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
Tse3
|
| Synonyms | |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | 2.97 |
| Kyte-Doolittle Hydrophobicity Value | -0.203 |
| Molecular Weight (kDa) | 44.4 |
| Isoelectric Point (pI) | 9.07 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3WA5 | HYDROLASE | 04/26/13 | Crystal Structure of type VI peptidoglycan muramidase effector Tse3 in complex with its cognate immunity protein Tsi3 | Pseudomonas aeruginosa | 1.9 | X-RAY DIFFRACTION | 100.0 |
| 4N80 | HYDROLASE/HYDROLASE INHIBITOR | 10/16/13 | Crystal structure of Tse3-Tsi3 complex | Pseudomonas aeruginosa | 2.4 | X-RAY DIFFRACTION | 100.0 |
| 4N7S | HYDROLASE/HYDROLASE INHIBITOR | 10/16/13 | Crystal structure of Tse3-Tsi3 complex with Zinc ion | Pseudomonas aeruginosa | 2.101 | X-RAY DIFFRACTION | 100.0 |
| 4N88 | HYDROLASE/HYDROLASE INHIBITOR | 10/17/13 | Crystal structure of Tse3-Tsi3 complex with calcium ion | Pseudomonas aeruginosa | 2.8 | X-RAY DIFFRACTION | 100.0 |
| 4M5E | HYDROLASE | 08/08/13 | Tse3 structure | Pseudomonas aeruginosa | 1.49 | X-RAY DIFFRACTION | 100.0 |
| 4M5F | HYDROLASE INHIBITOR | 08/08/13 | complex structure of Tse3-Tsi3 | Pseudomonas aeruginosa | 2.5 | X-RAY DIFFRACTION | 100.0 |
| 4LUQ | PROTEIN BINDING/TOXIN INHIBITOR | 07/25/13 | Crystal structure of virulence effector Tse3 in complex with neutralizer Tsi3 | Pseudomonas aeruginosa | 1.77 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 2 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG002158 (298 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3484
Search term: tse3
Search term: Tse3
|
Human Homologs
References
|
Cloning and characterization of the MamI restriction-modification system from Microbacterium ammoniaphilum in Escherichia coli.
Striebel HM, Seeber S, Jarsch M, Kessler C
Gene 1996 Jun 12;172(1):41-6
PubMed ID: 8654988
|
|
A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria.
Hood RD, Singh P, Hsu F, Güvener T, Carl MA, Trinidad RR, Silverman JM, Ohlson BB, Hicks KG, Plemel RL, Li M, Schwarz S, Wang WY, Merz AJ, Goodlett DR, Mougous JD
Cell Host Microbe 2010 Jan 21;7(1):25-37
PubMed ID: 20114026
|
|
Type VI secretion delivers bacteriolytic effectors to target cells.
Russell AB, Hood RD, Bui NK, LeRoux M, Vollmer W, Mougous JD
Nature 2011 Jul 20;475(7356):343-7
PubMed ID: 21776080
|