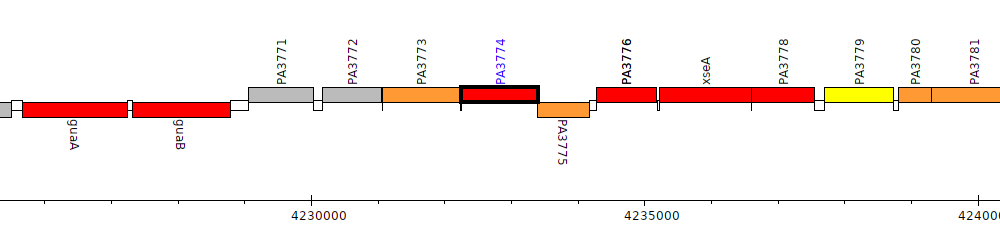
Pseudomonas aeruginosa PAO1, PA3774
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA3774
|
| Name |
|
| Replicon | chromosome |
| Genomic location | 4232258 - 4233400 (+ strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252463.1 |
| GI | 15598969 |
| Affymetrix | PA3774_at |
| Entrez | 880599 |
| GenBank | AAG07161.1 |
| INSDC | AAG07161.1 |
| NCBI Locus Tag | PA3774 |
| protein_id(GenBank) | gb|AAG07161.1|AE004796_6|gnl|PseudoCAP|PA3774 |
| TIGR | NTL03PA03774 |
| UniParc | UPI00000C5B06 |
| UniProtKB Acc | Q9HXM1 |
| UniProtKB ID | Q9HXM1_PSEAE |
| UniRef100 | UniRef100_S0J5Y1 |
| UniRef50 | UniRef50_S0ACH9 |
| UniRef90 | UniRef90_V6AAR1 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
histone deacetylase-like amidohydrolase
|
| Synonyms |
probable acetylpolyamine aminohydrolase |
| Evidence for Translation | |
| Charge (pH 7) | -5.57 |
| Kyte-Doolittle Hydrophobicity Value | -0.113 |
| Molecular Weight (kDa) | 41.0 |
| Isoelectric Point (pI) | 6.34 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 5G13 | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH (H143A) unliganded. | PSEUDOMONAS AERUGINOSA | 1.99 | X-RAY DIFFRACTION | 99.7 |
| 5G0Y | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH unliganded. | PSEUDOMONAS AERUGINOSA | 2.29 | X-RAY DIFFRACTION | 100.0 |
| 5G10 | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH bound to 9,9,9 trifluoro-8,8-dihydroy-N-phenylnonanamide | PSEUDOMONAS AERUGINOSA | 1.71 | X-RAY DIFFRACTION | 100.0 |
| 5G12 | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH (Y313F) unliganded. | PSEUDOMONAS AERUGINOSA | 2.02 | X-RAY DIFFRACTION | 99.7 |
| 5LI3 | SIGNALING PROTEIN | 07/14/16 | Crystal structure of HDAC-like protein from P. aeruginosa in complex with a photo-switchable inhibitor. | Pseudomonas aeruginosa | 2.4 | X-RAY DIFFRACTION | 100.0 |
| 5G0X | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH bound to acetate. | PSEUDOMONAS AERUGINOSA | 1.7 | X-RAY DIFFRACTION | 100.0 |
| 5G11 | HYDROLASE | 03/23/16 | Pseudomonas aeruginosa HDAH bound to PFSAHA. | PSEUDOMONAS AERUGINOSA | 2.48 | X-RAY DIFFRACTION | 99.7 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 443 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001890 (353 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA3774
Search term: histone deacetylase-like amidohydrolase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
histone deacetylase 10 [Source:HGNC Symbol;Acc:HGNC:18128]
E-value:
1.7e-39
Percent Identity:
37.3
|
References
|
A fluorescence lifetime-based binding assay for acetylpolyamine amidohydrolases from Pseudomonas aeruginosa using a [1,3]dioxolo[4,5-f][1,3]benzodioxole (DBD) ligand probe.
Meyners C, Wawrzinek R, Krämer A, Hinz S, Wessig P, Meyer-Almes FJ
Anal Bioanal Chem 2014 Aug;406(20):4889-97
PubMed ID: 24871864
|
|
Substrate specificity and function of acetylpolyamine amidohydrolases from Pseudomonas aeruginosa.
Krämer A, Herzer J, Overhage J, Meyer-Almes FJ
BMC Biochem 2016 Mar 9;17:4
PubMed ID: 26956223
|