Pseudomonas aeruginosa PAO1, PA4043 (ispA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
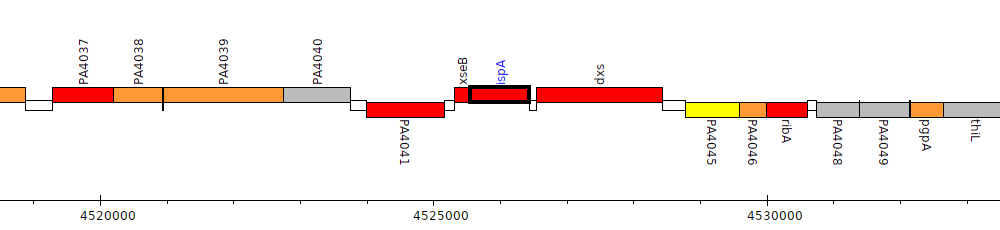
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4043
|
| Name |
ispA
|
| Replicon | chromosome |
| Genomic location | 4525551 - 4526438 (+ strand) |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_252732.1 |
| GI | 15599238 |
| Affymetrix | PA4043_ispA_at |
| DNASU | PaCD00007766 |
| Entrez | 879058 |
| GenBank | AAG07430.1 |
| INSDC | AAG07430.1 |
| NCBI Locus Tag | PA4043 |
| protein_id(GenBank) | gb|AAG07430.1|AE004821_3|gnl|PseudoCAP|PA4043 |
| TIGR | NTL03PA04043 |
| UniParc | UPI00000C5BD9 |
| UniProtKB Acc | Q9HWY4 |
| UniProtKB ID | Q9HWY4_PSEAE |
| UniRef100 | UniRef100_Q9HWY4 |
| UniRef50 | UniRef50_A0A011QRS6 |
| UniRef90 | UniRef90_Q9HWY4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
geranyltranstransferase
|
| Synonyms |
farnesyldiphosphate synthase FPP synthase |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -8.43 |
| Kyte-Doolittle Hydrophobicity Value | -0.046 |
| Molecular Weight (kDa) | 31.5 |
| Isoelectric Point (pI) | 5.06 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3ZOU | TRANSFERASE | 02/25/13 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696, and substrate geranyl pyrophosphate. | PSEUDOMONAS AERUGINOSA PAO1 | 1.55 | X-RAY DIFFRACTION | 100.0 |
| 3ZL6 | TRANSFERASE | 01/28/13 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PAO1, with bound fragment KM10833. | PSEUDOMONAS AERUGINOSA PAO1 | 1.85 | X-RAY DIFFRACTION | 100.0 |
| 4UMJ | TRANSFERASE | 05/18/14 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound ibandronic acid molecules. | PSEUDOMONAS AERUGINOSA | 1.85 | X-RAY DIFFRACTION | 100.0 |
| 3ZMC | TRANSFERASE | 02/07/13 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound substrate molecule Geranyl pyrophosphate. | PSEUDOMONAS AERUGINOSA PAO1 | 1.87 | X-RAY DIFFRACTION | 100.0 |
| 3ZMB | TRANSFERASE | 02/07/13 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01, with bound fragment SPB02696. | PSEUDOMONAS AERUGINOSA PAO1 | 1.9 | X-RAY DIFFRACTION | 100.0 |
| 3ZCD | TRANSFERASE | 11/19/12 | Native structure of Farnesyl Pyrophosphate Synthase from Pseudomonas aeruginosa PA01. | PSEUDOMONAS AERUGINOSA PAO1 | 1.55 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 656 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001641 (538 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4043
Search term: ispA
Search term: geranyltranstransferase
|
Human Homologs
References
|
Cloning and nucleotide sequence of the ispA gene responsible for farnesyl diphosphate synthase activity in Escherichia coli.
Fujisaki S, Hara H, Nishimura Y, Horiuchi K, Nishino T
J Biochem 1990 Dec;108(6):995-1000
PubMed ID: 2089044
|
|
The complete genome sequence of Escherichia coli K-12.
Blattner FR, Plunkett G 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, Gregor J, Davis NW, Kirkpatrick HA, Goeden MA, Rose DJ, Mau B, Shao Y
Science 1997 Sep 5;277(5331):1453-62
PubMed ID: 9278503
|
|
Cloning and characterization of a gene from Escherichia coli encoding a transketolase-like enzyme that catalyzes the synthesis of D-1-deoxyxylulose 5-phosphate, a common precursor for isoprenoid, thiamin, and pyridoxol biosynthesis.
Lois LM, Campos N, Putra SR, Danielsen K, Rohmer M, Boronat A
Proc Natl Acad Sci U S A 1998 Mar 3;95(5):2105-10
PubMed ID: 9482846
|