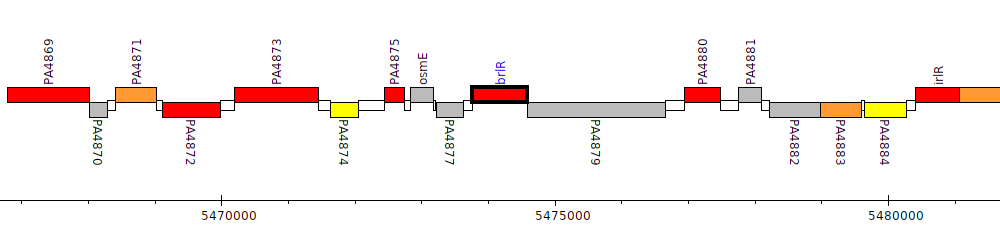
Pseudomonas aeruginosa PAO1, PA4878 (brlR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4878
|
| Name |
brlR
|
| Replicon | chromosome |
| Genomic location | 5473766 - 5474578 (+ strand) |
| Transposon Mutants | 2 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_253565.1 |
| GI | 15600071 |
| Affymetrix | PA4878_at |
| DNASU | PaCD00008112 |
| Entrez | 882234 |
| GenBank | AAG08263.1 |
| INSDC | AAG08263.1 |
| NCBI Locus Tag | PA4878 |
| protein_id(GenBank) | gb|AAG08263.1|AE004901_5|gnl|PseudoCAP|PA4878 |
| TIGR | NTL03PA04879 |
| UniParc | UPI00000C5E54 |
| UniProtKB Acc | Q9HUT5 |
| UniProtKB ID | Q9HUT5_PSEAE |
| UniRef100 | UniRef100_W0YYW8 |
| UniRef50 | UniRef50_A6VCY6 |
| UniRef90 | UniRef90_W0YYW8 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
BrlR
|
| Synonyms | |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -0.39 |
| Kyte-Doolittle Hydrophobicity Value | -0.451 |
| Molecular Weight (kDa) | 31.2 |
| Isoelectric Point (pI) | 6.92 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 5XBW | DNA BINDING PROTEIN | 03/21/17 | The structure of BrlR | Pseudomonas aeruginosa PAO1 | 3.109 | X-RAY DIFFRACTION | 100.0 |
| 5XBT | DNA BINDING PROTEIN | 03/21/17 | The structure of BrlR bound to c-di-GMP | Pseudomonas aeruginosa PAO1 | 2.495 | X-RAY DIFFRACTION | 100.0 |
| 5YC9 | TRANSCRIPTION | 09/07/17 | Crystal structure of a Pseudomonas aeruginosa transcriptional regulator | Pseudomonas aeruginosa | 2.9 | X-RAY DIFFRACTION | 100.0 |
| 5XBI | TRANSCRIPTION | 03/17/17 | The structure of BrlR-C domain bound to 3-amino-2-phenazino(a pyocyanin analog) | Pseudomonas aeruginosa | 1.4 | X-RAY DIFFRACTION | 99.3 |
| 5XQL | TRANSCRIPTION | 06/07/17 | Crystal structure of a Pseudomonas aeruginosa transcriptional regulator | Pseudomonas aeruginosa | 2.494 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 235 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG004506 (322 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4878
Search term: brlR
Search term: BrlR
|
Human Homologs
References
|
The MerR-like transcriptional regulator BrlR contributes to Pseudomonas aeruginosa biofilm tolerance.
Liao J, Sauer K
J Bacteriol 2012 Sep;194(18):4823-36
PubMed ID: 22730129
|
|
The MerR-like regulator BrlR confers biofilm tolerance by activating multidrug efflux pumps in Pseudomonas aeruginosa biofilms.
Liao J, Schurr MJ, Sauer K
J Bacteriol 2013 Aug;195(15):3352-63
PubMed ID: 23687276
|
|
The MerR-like regulator BrlR impairs Pseudomonas aeruginosa biofilm tolerance to colistin by repressing PhoPQ.
Chambers JR, Sauer K
J Bacteriol 2013 Oct;195(20):4678-88
PubMed ID: 23935054
|