Pseudomonas aeruginosa PAO1, PA4976 (aruH)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
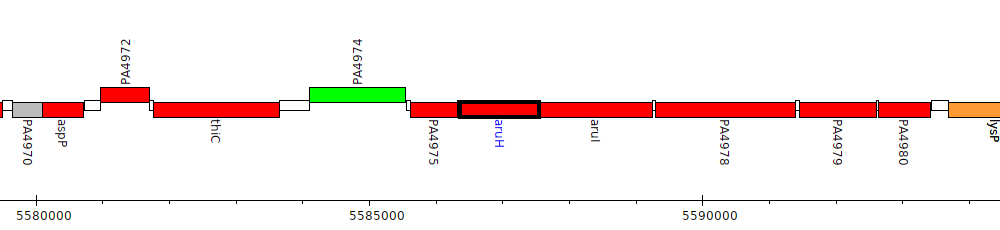
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA4976
|
| Name |
aruH
Synonym: aruH |
| Replicon | chromosome |
| Genomic location | 5586370 - 5587551 (- strand) |
| Transposon Mutants | 4 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 1 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_253663.1 |
| GI | 15600169 |
| Affymetrix | PA4976_aspC_at |
| Entrez | 880915 |
| GenBank | AAG08361.1 |
| INSDC | AAG08361.1 |
| NCBI Locus Tag | PA4976 |
| protein_id(GenBank) | gb|AAG08361.1|AE004910_8|gnl|PseudoCAP|PA4976 |
| TIGR | NTL03PA04977 |
| UniParc | UPI00000C5EA5 |
| UniProtKB Acc | Q9HUI9 |
| UniProtKB ID | ARUH_PSEAE |
| UniRef100 | UniRef100_Q9HUI9 |
| UniRef50 | UniRef50_Q9HUI9 |
| UniRef90 | UniRef90_Q9HUI9 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
Arginine:Pyruvate Transaminas, AruH
|
| Synonyms |
ASPAT |
| Evidence for Translation | |
| Charge (pH 7) | -9.89 |
| Kyte-Doolittle Hydrophobicity Value | 0.072 |
| Molecular Weight (kDa) | 42.4 |
| Isoelectric Point (pI) | 5.27 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 652 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG004590 (439 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA4976
Search term: aruH
Search term: Arginine:Pyruvate Transaminas, AruH
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
kynurenine aminotransferase 3 [Source:HGNC Symbol;Acc:HGNC:33238]
E-value:
7.2e-38
Percent Identity:
30.2
|
|
Ensembl
110, assembly
GRCh38.p14
|
kynurenine aminotransferase 3 [Source:HGNC Symbol;Acc:HGNC:33238]
E-value:
7.2e-38
Percent Identity:
30.2
|
References
|
Cloning and nucleotide sequencing of Rhizobium meliloti aminotransferase genes: an aspartate aminotransferase required for symbiotic nitrogen fixation is atypical.
Watson RJ, Rastogi VK
J Bacteriol 1993 Apr;175(7):1919-28
PubMed ID: 8096210
|
|
Stereospecific production of the herbicide phosphinothricin (glufosinate): purification of aspartate transaminase from Bacillus stearothermophilus, cloning of the corresponding gene, aspC, and application in a coupled transaminase process.
Bartsch K, Schneider R, Schulz A
Appl Environ Microbiol 1996 Oct;62(10):3794-9
PubMed ID: 8837436
|
|
Functional genomics enables identification of genes of the arginine transaminase pathway in Pseudomonas aeruginosa.
Yang Z, Lu CD
J Bacteriol 2007 Jun;189(11):3945-53
PubMed ID: 17416670
|
|
Characterization of an arginine:pyruvate transaminase in arginine catabolism of Pseudomonas aeruginosa PAO1.
Yang Z, Lu CD
J Bacteriol 2007 Jun;189(11):3954-9
PubMed ID: 17416668
|