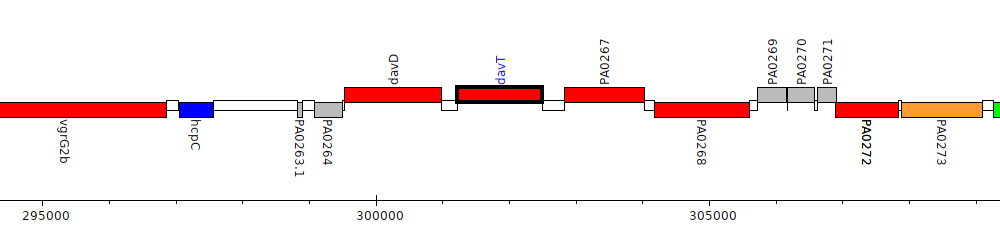
Pseudomonas aeruginosa PAO1, PA0266 (davT)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA0266
|
| Name |
davT
Synonym: gabT |
| Replicon | chromosome |
| Genomic location | 301218 - 302498 (+ strand) |
| Transposon Mutants | 3 transposon mutants in PAO1 |
| Transposon Mutants in orthologs | 3 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_248957.1 |
| GI | 15595463 |
| Affymetrix | PA0266_gabT_at |
| Entrez | 880917 |
| GenBank | AAG03655.1 |
| INSDC | AAG03655.1 |
| NCBI Locus Tag | PA0266 |
| protein_id(GenBank) | gb|AAG03655.1|AE004465_1|gnl|PseudoCAP|PA0266 |
| TIGR | NTL03PA00267 |
| UniParc | UPI00000C4FCE |
| UniProtKB Acc | Q9I6M4 |
| UniProtKB ID | DAVT_PSEAE |
| UniRef100 | UniRef100_Q9I6M4 |
| UniRef50 | UniRef50_P50457 |
| UniRef90 | UniRef90_Q9I6M4 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
delta-aminovalerate aminotransferase
|
| Synonyms |
4-aminobutyrate aminotransferase gabA transaminase |
| Evidence for Translation |
Detected by MALDI-TOF/TOF (PMID:19333994).
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -3.00 |
| Kyte-Doolittle Hydrophobicity Value | 0.142 |
| Molecular Weight (kDa) | 45.2 |
| Isoelectric Point (pI) | 6.43 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 639 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG000263 (534 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA0266
Search term: davT
Search term: delta-aminovalerate aminotransferase
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
ornithine aminotransferase [Source:HGNC Symbol;Acc:HGNC:8091]
E-value:
2.4e-47
Percent Identity:
32.1
|
References
|
Stereospecific production of the herbicide phosphinothricin (glufosinate) by transamination: isolation and characterization of a phosphinothricin-specific transaminase from Escherichia coli.
Schulz A, Taggeselle P, Tripier D, Bartsch K
Appl Environ Microbiol 1990 Jan;56(1):1-6
PubMed ID: 2178550
|
|
Molecular analysis of two genes of the Escherichia coli gab cluster: nucleotide sequence of the glutamate:succinic semialdehyde transaminase gene (gabT) and characterization of the succinic semialdehyde dehydrogenase gene (gabD).
Bartsch K, von Johnn-Marteville A, Schulz A
J Bacteriol 1990 Dec;172(12):7035-42
PubMed ID: 2254272
|
|
Functional characterization of seven γ-Glutamylpolyamine synthetase genes and the bauRABCD locus for polyamine and β-Alanine utilization in Pseudomonas aeruginosa PAO1.
Yao X, He W, Lu CD
J. Bacteriol. 2011 Aug;193(15):3923-30
PubMed ID: 21622750
|
|
Analysis of the periplasmic proteome of Pseudomonas aeruginosa, a metabolically versatile opportunistic pathogen.
Imperi F, Ciccosanti F, Perdomo AB, Tiburzi F, Mancone C, Alonzi T, Ascenzi P, Piacentini M, Visca P, Fimia GM
Proteomics 2009 Apr;9(7):1901-15
PubMed ID: 19333994
|
|
Multiple and interconnected pathways for L-lysine catabolism in Pseudomonas putida KT2440.
Revelles O, Espinosa-Urgel M, Fuhrer T, Sauer U, Ramos JL
J Bacteriol 2005 Nov;187(21):7500-10
PubMed ID: 16237033
|