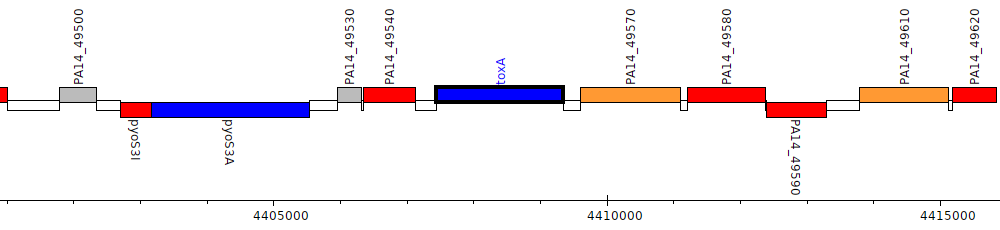
Pseudomonas aeruginosa UCBPP-PA14, PA14_49560 (toxA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa UCBPP-PA14 (Lee et al., 2006)
GCF_000014625.1|latest |
| Locus Tag |
PA14_49560
|
| Name |
toxA
|
| Replicon | chromosome |
| Genomic location | 4407441 - 4409348 (+ strand) |
| Transposon Mutants | 2 transposon mutants in UCBPP-PA14 |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | YP_792118.1 |
| GI | 116049080 |
| Entrez | 4381678 |
| INSDC | ABJ10316.1 |
| NCBI Locus Tag | PA14_49560 |
| UniParc | UPI000045D0C3 |
| UniProtKB Acc | Q02IW8 |
| UniProtKB ID | Q02IW8_PSEAB |
| UniRef100 | UniRef100_Q02IW8 |
| UniRef50 | UniRef50_P11439 |
| UniRef90 | UniRef90_P11439 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
exotoxin A
|
| Synonyms | |
| Evidence for Translation | |
| Charge (pH 7) | -17.12 |
| Kyte-Doolittle Hydrophobicity Value | -0.263 |
| Molecular Weight (kDa) | 69.1 |
| Isoelectric Point (pI) | 5.21 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 1DMA | ADP-RIBOSYLATION | 04/28/95 | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH NICOTINAMIDE AND AMP | Pseudomonas aeruginosa | 2.5 | X-RAY DIFFRACTION | 98.6 |
| 1ZM4 | BIOSYNTHETIC PROTEIN/TRANSFERASE | 05/10/05 | Structure of the eEF2-ETA-bTAD complex | Pseudomonas aeruginosa; Saccharomyces cerevisiae | 2.9 | X-RAY DIFFRACTION | 99.0 |
| 6EDG | TRANSFERASE | 08/09/18 | Pseudomonas exotoxin A domain III T18H477L | Pseudomonas aeruginosa | 1.47 | X-RAY DIFFRACTION | 96.7 |
| 1ZM3 | BIOSYNTHETIC PROTEIN/TRANSFERASE | 05/10/05 | Structure of the apo eEF2-ETA complex | Pseudomonas aeruginosa; Saccharomyces cerevisiae | 3.07 | X-RAY DIFFRACTION | 99.0 |
| 1XK9 | TRANSFERASE | 09/28/04 | Pseudomanas exotoxin A in complex with the PJ34 inhibitor | Pseudomonas aeruginosa | 2.1 | X-RAY DIFFRACTION | 98.6 |
| 2ZIT | BIOSYNTHETIC PROTEIN/TRANSFERASE | 02/24/08 | Structure of the eEF2-ExoA-NAD+ complex | Pseudomonas aeruginosa; Saccharomyces cerevisiae | 3 | X-RAY DIFFRACTION | 99.0 |
| 1IKQ | TRANSFERASE | 05/04/01 | Pseudomonas Aeruginosa Exotoxin A, wild type | Pseudomonas aeruginosa | 1.62 | X-RAY DIFFRACTION | 98.7 |
| 1ZM2 | BIOSYNTHETIC PROTEIN/TRANSFERASE | 05/10/05 | Structure of ADP-ribosylated eEF2 in complex with catalytic fragment of ETA | Pseudomonas aeruginosa; Saccharomyces cerevisiae | 3.07 | X-RAY DIFFRACTION | 99.0 |
| 1AER | ADP-RIBOSYLATION | 12/11/95 | DOMAIN III OF PSEUDOMONAS AERUGINOSA EXOTOXIN COMPLEXED WITH BETA-TAD | Pseudomonas aeruginosa | 2.3 | X-RAY DIFFRACTION | 98.6 |
| 1IKP | TRANSFERASE | 05/04/01 | Pseudomonas Aeruginosa Exotoxin A, P201Q, W281A mutant | Pseudomonas aeruginosa | 1.45 | X-RAY DIFFRACTION | 98.4 |
| 1ZM9 | BIOSYNTHETIC PROTEIN/TRANSFERASE | 05/10/05 | Structure of eEF2-ETA in complex with PJ34 | Pseudomonas aeruginosa; Saccharomyces cerevisiae | 2.8 | X-RAY DIFFRACTION | 99.0 |
Virulence Evidence
| Source 1 | |
| Database | PseudoCAP |
| Category | Exototoxin A |
| Evidence |
ECO:0000314
direct assay evidence used in manual assertion
|
| Host Organism | Mus musculus |
| Notes | |
| Infection model | Burned mouse model |
| Reference | 10922040 |
| Source 2 | |
| Database | Victors : 4410 |
| Category | Exototoxin A |
| Evidence |
ECO:0000015
mutant phenotype evidence
|
| Host Organism | Mus musculus |
| Notes | |
| Reference | 7604262 |
| Source 3 | |
| Database | PseudoCAP |
| Category | Exototoxin A |
| Evidence |
ECO:0000314
direct assay evidence used in manual assertion
|
| Host Organism | Arabidopsis thaliana |
| Notes | |
| Infection model | Arabidopsis leaf infiltration assay |
| Reference | 10922040 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in one genus
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG001108 (295 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA14_49560
Search term: toxA
Search term: exotoxin A
|
Human Homologs
References
No references are associated with this feature.