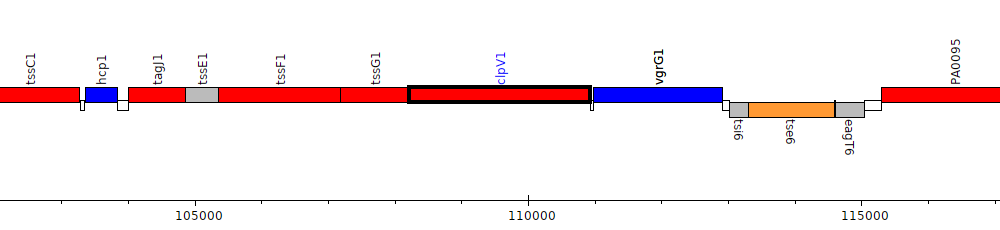
Pseudomonas aeruginosa PAO1, PA0090 (clpV1)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SM00382
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF07724
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02025 | Biofilm formation - Pseudomonas aeruginosa | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae03070 | Bacterial secretion system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 743 | 757 | 4.6E-30 |
| Gene3D | G3DSA:1.10.1780.10 | - | IPR036628 | Clp, N-terminal domain superfamily | 1 | 161 | 5.2E-41 |
| SMART | SM01086 | ClpB_D2_small_2 | IPR019489 | Clp ATPase, C-terminal | 804 | 896 | 5.4E-11 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 229 | 370 | 4.8E-10 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 581 | 878 | 5.57E-63 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 585 | 802 | 2.3E-66 |
| PANTHER | PTHR11638 | ATP-DEPENDENT CLP PROTEASE | - | - | 18 | 877 | 0.0 |
| SUPERFAMILY | SSF81923 | Double Clp-N motif | IPR036628 | Clp, N-terminal domain superfamily | 12 | 133 | 1.41E-17 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 636 | 654 | 4.6E-30 |
| FunFam | G3DSA:3.40.50.300:FF:000010 | Chaperone clpB 1, putative | - | - | 182 | 373 | 2.5E-83 |
| Coils | Coil | Coil | - | - | 443 | 463 | - |
| Pfam | PF07724 | AAA domain (Cdc48 subfamily) | IPR003959 | ATPase, AAA-type, core | 631 | 797 | 2.0E-38 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 710 | 728 | 4.6E-30 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 375 | 579 | 5.8E-54 |
| Pfam | PF10431 | C-terminal, D2-small domain, of ClpB protein | IPR019489 | Clp ATPase, C-terminal | 804 | 878 | 1.8E-12 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 632 | 775 | 2.5E-9 |
| NCBIfam | TIGR03345 | JCVI: type VI secretion system ATPase TssH | IPR017729 | AAA+ ATPase ClpV1 | 13 | 888 | 0.0 |
| Pfam | PF00004 | ATPase family associated with various cellular activities (AAA) | IPR003959 | ATPase, AAA-type, core | 234 | 367 | 2.5E-9 |
| Gene3D | G3DSA:1.10.8.60 | - | - | - | 804 | 900 | 3.1E-7 |
| Coils | Coil | Coil | - | - | 474 | 508 | - |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 183 | 373 | 8.4E-72 |
| CDD | cd19499 | RecA-like_ClpB_Hsp104-like | - | - | 594 | 802 | 5.2044E-91 |
| FunFam | G3DSA:3.40.50.300:FF:000025 | ATP-dependent Clp protease subunit | - | - | 585 | 802 | 2.4E-78 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 190 | 519 | 9.83E-73 |
| PRINTS | PR00300 | ATP-dependent Clp protease ATP-binding subunit signature | IPR001270 | ClpA/B family | 681 | 699 | 4.6E-30 |
| Coils | Coil | Coil | - | - | 531 | 558 | - |
| CDD | cd00009 | AAA | - | - | 212 | 367 | 4.58994E-15 |
| Pfam | PF17871 | AAA lid domain | IPR041546 | ClpA/ClpB, AAA lid domain | 373 | 468 | 3.9E-28 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.