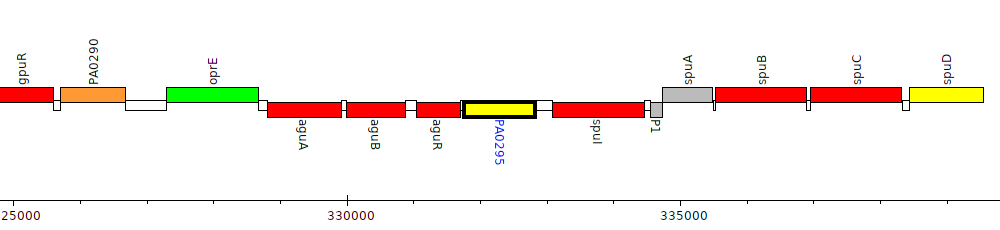
Pseudomonas aeruginosa PAO1, PA0295
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0019810 | putrescine binding |
Inferred from Sequence or Structural Similarity
Term mapped from: PDB:1A99
|
ECO:0007090 |
31631799 | Reviewed by curator |
| Cellular Component | GO:0042597 | periplasmic space |
Inferred from Sequence Model
Term mapped from: InterPro:PR00909
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0019808 | polyamine binding |
Inferred from Sequence Model
Term mapped from: InterPro:PR00909
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0015846 | polyamine transport |
Inferred from Sequence Model
Term mapped from: InterPro:PR00909
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF13416 | Bacterial extracellular solute-binding protein | IPR006059 | Bacterial extracellular solute-binding protein | 36 | 310 | 2.9E-27 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 147 | 161 | 1.1E-38 |
| SUPERFAMILY | SSF53850 | Periplasmic binding protein-like II | - | - | 20 | 348 | 3.23E-78 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 117 | 133 | 1.1E-38 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 296 | 322 | 1.1E-38 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 26 | 42 | 1.1E-38 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 170 | 189 | 1.1E-38 |
| PANTHER | PTHR30222 | SPERMIDINE/PUTRESCINE-BINDING PERIPLASMIC PROTEIN | - | - | 17 | 348 | 4.4E-73 |
| Gene3D | G3DSA:3.40.190.10 | - | - | - | 23 | 318 | 2.6E-107 |
| CDD | cd13659 | PBP2_PotF | - | - | 22 | 345 | 0.0 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 251 | 270 | 1.1E-38 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 42 | 63 | 1.1E-38 |
| Gene3D | G3DSA:3.40.190.10 | - | - | - | 126 | 346 | 2.6E-107 |
| PIRSF | PIRSF019574 | Periplasmic_polyamine_BP | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 4 | 352 | 1.3E-97 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 207 | 226 | 1.1E-38 |
| PRINTS | PR00909 | Bacterial periplasmic spermidine/putrescine-binding protein signature | IPR001188 | Spermidine/putrescine-binding periplasmic protein | 84 | 97 | 1.1E-38 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.