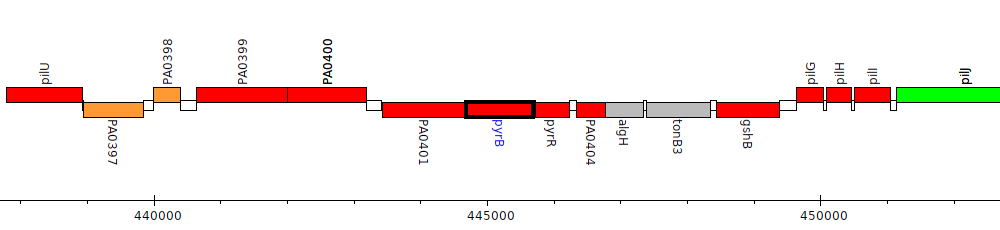
Pseudomonas aeruginosa PAO1, PA0402 (pyrB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006531 | aspartate metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0009117 | nucleotide metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006520 | cellular amino acid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006522 | alanine metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0004355 | glutamate synthase (NADPH) activity | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006220 | pyrimidine nucleotide metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0016597 | amino acid binding |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53671
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004070 | aspartate carbamoyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00001
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016743 | carboxyl- or carbamoyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53671
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006520 | cellular amino acid metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53671
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0006207 | 'de novo' pyrimidine nucleobase biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00001
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Pyrimidine metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCyc | PWY-5686 | UMP biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCAP | Alanine and Aspartate metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCyc | PRPP-PWY | superpathway of histidine, purine, and pyrimidine biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00250 | Alanine, aspartate and glutamate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY0-162 | superpathway of pyrimidine ribonucleotides de novo biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00240 | Pyrimidine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR45753 | ORNITHINE CARBAMOYLTRANSFERASE, MITOCHONDRIAL | - | - | 14 | 319 | 4.9E-65 |
| PRINTS | PR00100 | Aspartate/ornithine carbamoyltransferase superfamily signature | IPR006130 | Aspartate/ornithine carbamoyltransferase | 284 | 307 | 7.7E-18 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 149 | 166 | 3.4E-31 |
| PRINTS | PR00100 | Aspartate/ornithine carbamoyltransferase superfamily signature | IPR006130 | Aspartate/ornithine carbamoyltransferase | 150 | 161 | 7.7E-18 |
| PRINTS | PR00100 | Aspartate/ornithine carbamoyltransferase superfamily signature | IPR006130 | Aspartate/ornithine carbamoyltransferase | 64 | 83 | 7.7E-18 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 92 | 101 | 3.4E-31 |
| Pfam | PF00185 | Aspartate/ornithine carbamoyltransferase, Asp/Orn binding domain | IPR006131 | Aspartate/ornithine carbamoyltransferase, Asp/Orn-binding domain | 170 | 317 | 1.9E-23 |
| Gene3D | G3DSA:3.40.50.1370 | Aspartate/ornithine carbamoyltransferase | IPR036901 | Aspartate/ornithine carbamoyltransferase superfamily | 19 | 315 | 5.6E-84 |
| Hamap | MF_00001 | Aspartate carbamoyltransferase [pyrB]. | IPR002082 | Aspartate carbamoyltransferase | 17 | 321 | 41.801838 |
| FunFam | G3DSA:3.40.50.1370:FF:000007 | Aspartate carbamoyltransferase | - | - | 153 | 306 | 1.6E-68 |
| NCBIfam | TIGR00670 | JCVI: aspartate carbamoyltransferase | IPR002082 | Aspartate carbamoyltransferase | 18 | 319 | 1.8E-90 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 54 | 76 | 3.4E-31 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 300 | 314 | 3.4E-31 |
| Pfam | PF02729 | Aspartate/ornithine carbamoyltransferase, carbamoyl-P binding domain | IPR006132 | Aspartate/ornithine carbamoyltransferase, carbamoyl-P binding | 18 | 162 | 2.8E-41 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 276 | 281 | 3.4E-31 |
| SUPERFAMILY | SSF53671 | Aspartate/ornithine carbamoyltransferase | IPR036901 | Aspartate/ornithine carbamoyltransferase superfamily | 17 | 321 | 3.8E-95 |
| PRINTS | PR00100 | Aspartate/ornithine carbamoyltransferase superfamily signature | IPR006130 | Aspartate/ornithine carbamoyltransferase | 272 | 281 | 7.7E-18 |
| FunFam | G3DSA:3.40.50.1370:FF:000006 | Aspartate carbamoyltransferase | - | - | 19 | 152 | 7.0E-73 |
| PRINTS | PR00101 | Aspartate carbamoyltransferase signature | - | - | 235 | 244 | 3.4E-31 |
| Gene3D | G3DSA:3.40.50.1370 | Aspartate/ornithine carbamoyltransferase | IPR036901 | Aspartate/ornithine carbamoyltransferase superfamily | 150 | 306 | 5.6E-84 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.