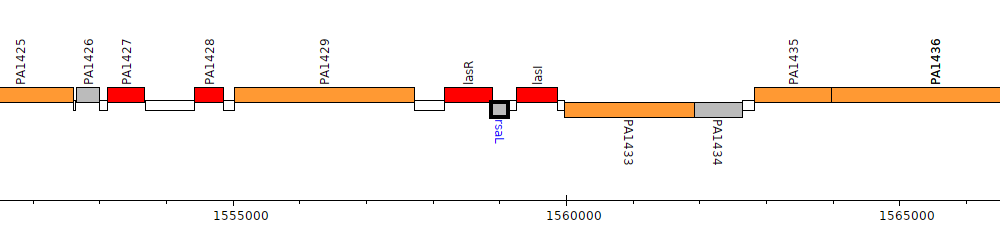
Pseudomonas aeruginosa PAO1, PA1431 (rsaL)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:1900377 | negative regulation of secondary metabolite biosynthetic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
18045385 | Reviewed by curator |
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
16385073 | Reviewed by curator |
| Biological Process | GO:1900192 | positive regulation of single-species biofilm formation | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19878323 | Reviewed by curator |
| Biological Process | GO:2000146 | negative regulation of cell motility | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19878323 | Reviewed by curator |
| Molecular Function | GO:0003677 | DNA binding | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
16385073 | Reviewed by curator |
| Biological Process | GO:0051713 | negative regulation of cytolysis in other organism | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19878323 | Reviewed by curator |
| Biological Process | GO:0009372 | quorum sensing | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
10094696 | Reviewed by curator |
| Biological Process | GO:0060311 | negative regulation of elastin catabolic process | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19878323 | Reviewed by curator |
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:SSF47413
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Quorum sensing |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF47413 | lambda repressor-like DNA-binding domains | IPR010982 | Lambda repressor-like, DNA-binding domain superfamily | 17 | 58 | 9.78E-5 |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 1 | 23 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 1 | 15 | - |
| Coils | Coil | Coil | - | - | 71 | 80 | - |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.