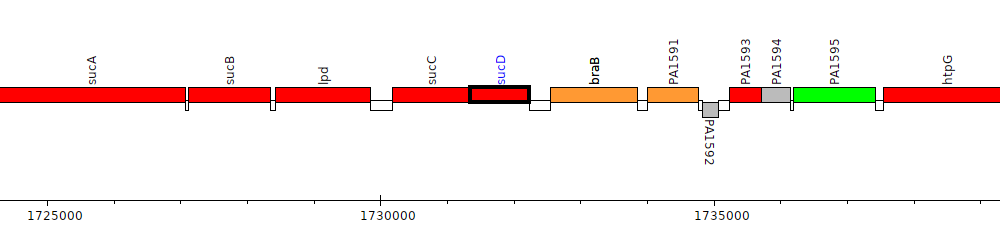
Pseudomonas aeruginosa PAO1, PA1589 (sucD)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009142 | nucleoside triphosphate biosynthetic process | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
10671455 | Reviewed by curator |
| Molecular Function | GO:0004550 | nucleoside diphosphate kinase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
10671455 | Reviewed by curator |
| Molecular Function | GO:0004775 | succinate-CoA ligase (ADP-forming) activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
10671455 | Reviewed by curator |
| Cellular Component | GO:0042709 | succinate-CoA ligase complex | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
10671455 | Reviewed by curator |
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF001553
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCyc | TCA | TCA cycle I (prokaryotic) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00640 | Propanoate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | GLYCOLYSIS-TCA-GLYOX-BYPASS | superpathway of glycolysis, pyruvate dehydrogenase, TCA, and glyoxylate bypass | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00660 | C5-Branched dibasic acid metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | GLUTAMOLYSIS-PWY | glutamate degradation III | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCAP | Citrate cycle (TCA cycle) |
ECO:0000037
not_recorded |
|||
| KEGG | pae00020 | Citrate cycle (TCA cycle) | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | P22-PWY | acetyl-CoA assimilation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | TCA-GLYOX-BYPASS | superpathway of glyoxylate bypass and TCA | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01200 | Carbon metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | C5-Branched dibasic acid metabolism |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR11117 | SUCCINYL-COA LIGASE SUBUNIT ALPHA | - | - | 3 | 287 | 0.0 |
| PRINTS | PR01798 | Succinyl-CoA synthase signature | - | - | 177 | 195 | 5.1E-39 |
| FunFam | G3DSA:3.40.50.720:FF:000002 | Succinate--CoA ligase [ADP-forming] subunit alpha | - | - | 1 | 120 | 3.0E-60 |
| PRINTS | PR01798 | Succinyl-CoA synthase signature | - | - | 240 | 257 | 5.1E-39 |
| PIRSF | PIRSF001553 | SucCS_alpha | IPR005810 | Succinyl-CoA ligase, alpha subunit | 1 | 290 | 0.0 |
| PRINTS | PR01798 | Succinyl-CoA synthase signature | - | - | 208 | 221 | 5.1E-39 |
| NCBIfam | TIGR01019 | JCVI: succinate--CoA ligase subunit alpha | IPR005810 | Succinyl-CoA ligase, alpha subunit | 4 | 287 | 0.0 |
| FunFam | G3DSA:3.40.50.261:FF:000002 | Succinate--CoA ligase [ADP-forming] subunit alpha | - | - | 120 | 290 | 3.6E-102 |
| SMART | SM00881 | CoA_binding_2 | IPR003781 | CoA-binding | 4 | 100 | 1.5E-36 |
| Gene3D | G3DSA:3.40.50.720 | - | - | - | 1 | 119 | 3.5E-56 |
| SUPERFAMILY | SSF52210 | Succinyl-CoA synthetase domains | IPR016102 | Succinyl-CoA synthetase-like | 123 | 289 | 1.16E-57 |
| Pfam | PF00549 | CoA-ligase | IPR005811 | ATP-citrate lyase/succinyl-CoA ligase | 151 | 270 | 6.5E-22 |
| Hamap | MF_01988 | Succinate--CoA ligase [ADP-forming] subunit alpha [sucD]. | IPR005810 | Succinyl-CoA ligase, alpha subunit | 1 | 288 | 42.831444 |
| PRINTS | PR01798 | Succinyl-CoA synthase signature | - | - | 82 | 99 | 5.1E-39 |
| Gene3D | G3DSA:3.40.50.261 | - | IPR016102 | Succinyl-CoA synthetase-like | 120 | 293 | 1.0E-77 |
| Pfam | PF02629 | CoA binding domain | IPR003781 | CoA-binding | 6 | 99 | 2.3E-31 |
| SUPERFAMILY | SSF51735 | NAD(P)-binding Rossmann-fold domains | IPR036291 | NAD(P)-binding domain superfamily | 1 | 122 | 1.93E-42 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.