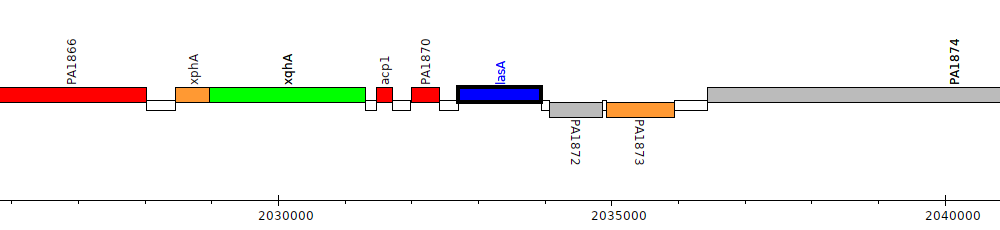
Pseudomonas aeruginosa PAO1, PA1871 (lasA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006508 | proteolysis | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
1597429 | Reviewed by curator |
| Biological Process | GO:0015628 | protein secretion by the type II secretion system | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
9642203 | Reviewed by curator |
| Molecular Function | GO:0004175 | endopeptidase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
9642203 | Reviewed by curator |
| Cellular Component | GO:0005615 | extracellular space | Inferred from Direct Assay | ECO:0001230 mass spectrometry evidence used in manual assertion |
25488299 | Reviewed by curator |
| Biological Process | GO:0043952 | protein transport by the Sec complex | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
9642203 | Reviewed by curator |
| Biological Process | GO:0006508 | proteolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PR00933
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004222 | metalloendopeptidase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR00933
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Xcp type II secretion system |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF01551 | Peptidase family M23 | IPR016047 | Peptidase M23 | 286 | 367 | 2.7E-11 |
| FunFam | G3DSA:2.70.70.10:FF:000044 | Protease LasA | - | - | 237 | 418 | 4.2E-129 |
| PANTHER | PTHR21666 | PEPTIDASE-RELATED | - | - | 205 | 368 | 2.3E-10 |
| Gene3D | G3DSA:2.70.70.10 | Glucose Permease (Domain IIA) | IPR011055 | Duplicated hybrid motif | 237 | 418 | 2.4E-54 |
| PRINTS | PR00933 | B-lytic metalloendopeptidase (M23) signature | IPR000841 | Peptidase M23A, B-lytic metalloendopeptidase | 244 | 265 | 1.6E-57 |
| PRINTS | PR00933 | B-lytic metalloendopeptidase (M23) signature | IPR000841 | Peptidase M23A, B-lytic metalloendopeptidase | 266 | 284 | 1.6E-57 |
| CDD | cd12797 | M23_peptidase | - | - | 287 | 361 | 6.00773E-17 |
| PRINTS | PR00933 | B-lytic metalloendopeptidase (M23) signature | IPR000841 | Peptidase M23A, B-lytic metalloendopeptidase | 372 | 395 | 1.6E-57 |
| SUPERFAMILY | SSF51261 | Duplicated hybrid motif | IPR011055 | Duplicated hybrid motif | 226 | 365 | 2.3E-17 |
| PRINTS | PR00933 | B-lytic metalloendopeptidase (M23) signature | IPR000841 | Peptidase M23A, B-lytic metalloendopeptidase | 293 | 311 | 1.6E-57 |
| PRINTS | PR00933 | B-lytic metalloendopeptidase (M23) signature | IPR000841 | Peptidase M23A, B-lytic metalloendopeptidase | 345 | 365 | 1.6E-57 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.