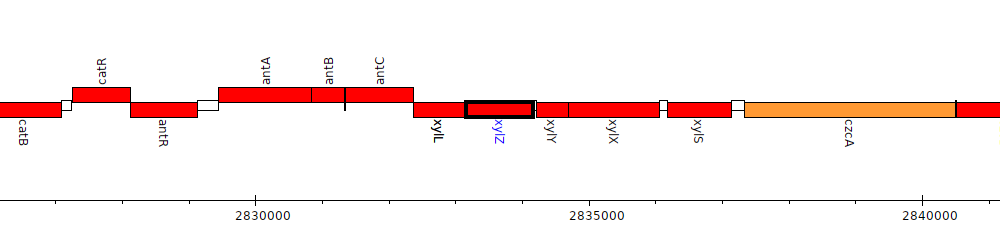
Pseudomonas aeruginosa PAO1, PA2516 (xylZ)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0044248 | cellular catabolic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
1938949 | Reviewed by curator |
| Molecular Function | GO:0018623 | benzoate 1,2-dioxygenase activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
1938949 | Reviewed by curator |
| Biological Process | GO:0006091 | generation of precursor metabolites and energy | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
1938949 | Reviewed by curator |
| Biological Process | GO:0015976 | carbon utilization | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00175
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0051536 | iron-sulfur cluster binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00111
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00364 | Fluorobenzoate degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01120 | Microbial metabolism in diverse environments | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Aromatic compound catabolism |
ECO:0000037
not_recorded |
|||
| KEGG | pae01220 | Degradation of aromatic compounds | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00622 | Xylene degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00362 | Benzoate degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF00111 | 2Fe-2S iron-sulfur cluster binding domain | IPR001041 | 2Fe-2S ferredoxin-type iron-sulfur binding domain | 10 | 85 | 4.8E-15 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 236 | 245 | 6.4E-25 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 153 | 160 | 6.4E-25 |
| PRINTS | PR00371 | Flavoprotein pyridine nucleotide cytochrome reductase signature | IPR001709 | Flavoprotein pyridine nucleotide cytochrome reductase | 300 | 308 | 5.4E-8 |
| CDD | cd00207 | fer2 | IPR001041 | 2Fe-2S ferredoxin-type iron-sulfur binding domain | 17 | 92 | 5.57376E-21 |
| Gene3D | G3DSA:3.40.50.80 | - | IPR039261 | Ferredoxin-NADP reductase (FNR), nucleotide-binding domain | 200 | 332 | 2.5E-35 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 196 | 205 | 6.4E-25 |
| SUPERFAMILY | SSF52343 | Ferredoxin reductase-like, C-terminal NADP-linked domain | IPR039261 | Ferredoxin-NADP reductase (FNR), nucleotide-binding domain | 204 | 333 | 3.67E-35 |
| PANTHER | PTHR47354 | NADH OXIDOREDUCTASE HCR | - | - | 17 | 91 | 1.4E-73 |
| SUPERFAMILY | SSF63380 | Riboflavin synthase domain-like | IPR017938 | Riboflavin synthase-like beta-barrel | 76 | 204 | 4.77E-23 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 136 | 148 | 6.4E-25 |
| SUPERFAMILY | SSF54292 | 2Fe-2S ferredoxin-like | IPR036010 | 2Fe-2S ferredoxin-like superfamily | 1 | 98 | 1.97E-25 |
| PRINTS | PR00371 | Flavoprotein pyridine nucleotide cytochrome reductase signature | IPR001709 | Flavoprotein pyridine nucleotide cytochrome reductase | 236 | 245 | 5.4E-8 |
| NCBIfam | NF040810 | NCBIFAM: benzoate 1,2-dioxygenase electron transfer component BenC | - | - | 3 | 335 | 0.0 |
| Gene3D | G3DSA:2.40.30.10 | Translation factors | - | - | 101 | 199 | 1.4E-22 |
| PRINTS | PR00371 | Flavoprotein pyridine nucleotide cytochrome reductase signature | IPR001709 | Flavoprotein pyridine nucleotide cytochrome reductase | 211 | 230 | 5.4E-8 |
| Gene3D | G3DSA:3.10.20.30 | - | IPR012675 | Beta-grasp domain superfamily | 1 | 100 | 2.9E-27 |
| PRINTS | PR00371 | Flavoprotein pyridine nucleotide cytochrome reductase signature | IPR001709 | Flavoprotein pyridine nucleotide cytochrome reductase | 153 | 160 | 5.4E-8 |
| Pfam | PF00970 | Oxidoreductase FAD-binding domain | IPR008333 | Flavoprotein pyridine nucleotide cytochrome reductase-like, FAD-binding domain | 109 | 202 | 5.9E-16 |
| CDD | cd06209 | BenDO_FAD_NAD | IPR047683 | Benzoate 1,2-dioxygenase reductase, FAD/NAD binding domain | 106 | 333 | 0.0 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 300 | 308 | 6.4E-25 |
| Pfam | PF00175 | Oxidoreductase NAD-binding domain | IPR001433 | Oxidoreductase FAD/NAD(P)-binding | 212 | 314 | 2.0E-22 |
| PRINTS | PR00410 | Phenol hydroxylase reductase family signature | - | - | 211 | 230 | 6.4E-25 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.