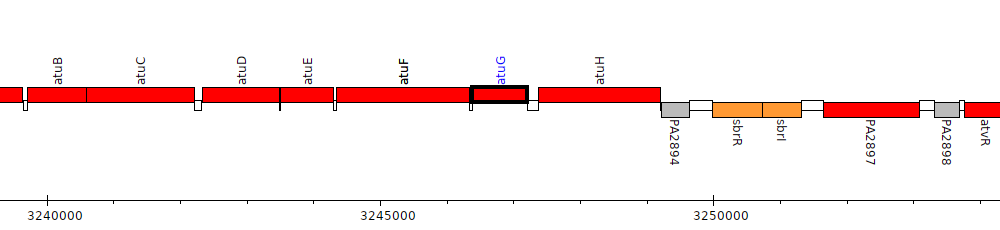
Pseudomonas aeruginosa PAO1, PA2892 (atuG)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0046247 | terpene catabolic process | Inferred from Genomic Context | |
18310025 | Reviewed by curator |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae00281 | Geraniol degradation | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.40.50.720:FF:000301 | Hydroxysteroid dehydrogenase like 2 | - | - | 1 | 274 | 6.2E-129 |
| Gene3D | G3DSA:3.40.50.720 | - | - | - | 1 | 274 | 5.4E-74 |
| PRINTS | PR00081 | Glucose/ribitol dehydrogenase family signature | IPR002347 | Short-chain dehydrogenase/reductase SDR | 135 | 151 | 1.8E-15 |
| SUPERFAMILY | SSF51735 | NAD(P)-binding Rossmann-fold domains | IPR036291 | NAD(P)-binding domain superfamily | 1 | 240 | 9.34E-53 |
| PRINTS | PR00081 | Glucose/ribitol dehydrogenase family signature | IPR002347 | Short-chain dehydrogenase/reductase SDR | 184 | 201 | 1.8E-15 |
| PANTHER | PTHR42808 | HYDROXYSTEROID DEHYDROGENASE-LIKE PROTEIN 2 | - | - | 2 | 274 | 1.0E-115 |
| Pfam | PF00106 | short chain dehydrogenase | IPR002347 | Short-chain dehydrogenase/reductase SDR | 6 | 196 | 4.5E-38 |
| PRINTS | PR00081 | Glucose/ribitol dehydrogenase family signature | IPR002347 | Short-chain dehydrogenase/reductase SDR | 163 | 182 | 1.8E-15 |
| PRINTS | PR00081 | Glucose/ribitol dehydrogenase family signature | IPR002347 | Short-chain dehydrogenase/reductase SDR | 7 | 24 | 1.8E-15 |
| PRINTS | PR00081 | Glucose/ribitol dehydrogenase family signature | IPR002347 | Short-chain dehydrogenase/reductase SDR | 88 | 99 | 1.8E-15 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.