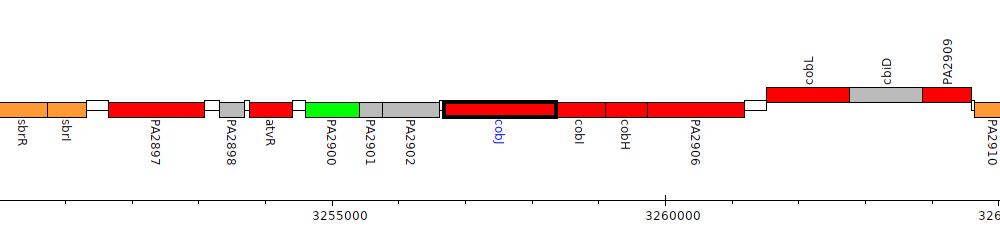
Pseudomonas aeruginosa PAO1, PA2903 (cobJ)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009236 | cobalamin biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
8226690 | Reviewed by curator |
| Biological Process | GO:0051188 | obsolete cofactor biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0008168 | methyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53790
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009236 | cobalamin biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:cd11646
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCyc | P381-PWY | adenosylcobalamin biosynthesis II (late cobalt incorporation) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00860 | Porphyrin and chlorophyll metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Cobalamin biosynthesis |
ECO:0000037
not_recorded |
|||
| PseudoCyc | PWY-7376 | cob(II)yrinate a,c-diamide biosynthesis II (late cobalt incorporation) | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| MetaCyc | cob(II)yrinate <i>a,c</i>-diamide biosynthesis I (early cobalt insertion) | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF159672 | CbiG N-terminal domain-like | IPR038029 | GbiG N-terminal domain superfamily | 7 | 212 | 1.18E-41 |
| Gene3D | G3DSA:3.40.1010.10 | - | IPR014777 | Tetrapyrrole methylase, subdomain 1 | 302 | 422 | 9.8E-27 |
| PANTHER | PTHR47036 | COBALT-FACTOR III C(17)-METHYLTRANSFERASE-RELATED | - | - | 300 | 558 | 1.1E-96 |
| SUPERFAMILY | SSF53790 | Tetrapyrrole methylase | IPR035996 | Tetrapyrrole methylase superfamily | 307 | 556 | 2.49E-53 |
| NCBIfam | TIGR01466 | JCVI: precorrin-3B C(17)-methyltransferase | IPR006363 | Precorrin-3B C17-methyltransferase domain | 310 | 556 | 1.4E-84 |
| Gene3D | G3DSA:3.40.50.11220 | - | - | - | 2 | 126 | 2.6E-20 |
| CDD | cd11646 | Precorrin_3B_C17_MT | IPR006363 | Precorrin-3B C17-methyltransferase domain | 310 | 556 | 9.05901E-114 |
| Pfam | PF00590 | Tetrapyrrole (Corrin/Porphyrin) Methylases | IPR000878 | Tetrapyrrole methylase | 309 | 520 | 4.5E-34 |
| Gene3D | G3DSA:3.30.950.10 | - | IPR014776 | Tetrapyrrole methylase, subdomain 2 | 423 | 557 | 9.9E-52 |
| Pfam | PF11760 | Cobalamin synthesis G N-terminal | IPR021744 | Cobalamin synthesis G, N-terminal | 52 | 130 | 7.3E-23 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.