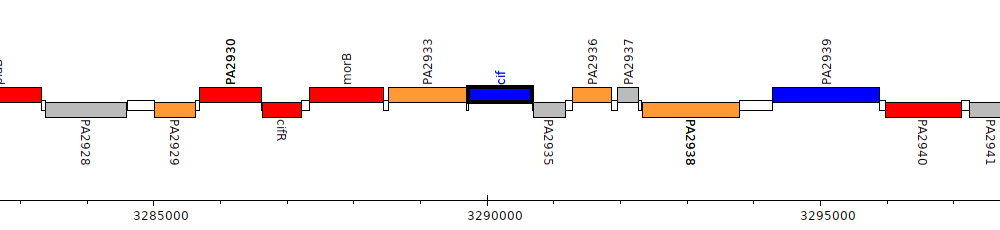
Pseudomonas aeruginosa PAO1, PA2934 (cif)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:2000157 | negative regulation of ubiquitin-specific protease activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
21455491 | Reviewed by curator |
| Molecular Function | GO:0004301 | epoxide hydrolase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
20118260 | Reviewed by curator |
| Biological Process | GO:0009405 | pathogenesis | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
16236828 | Reviewed by curator |
| Biological Process | GO:2001226 | negative regulation of chloride transport | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
16236828 | Reviewed by curator |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.1820 | alpha/beta hydrolase | IPR029058 | Alpha/Beta hydrolase fold | 33 | 316 | 2.2E-117 |
| Pfam | PF12697 | Alpha/beta hydrolase family | IPR000073 | Alpha/beta hydrolase fold-1 | 59 | 302 | 3.9E-18 |
| PANTHER | PTHR43329 | EPOXIDE HYDROLASE | - | - | 39 | 317 | 7.0E-33 |
| SUPERFAMILY | SSF53474 | alpha/beta-Hydrolases | IPR029058 | Alpha/Beta hydrolase fold | 31 | 317 | 1.62E-63 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.