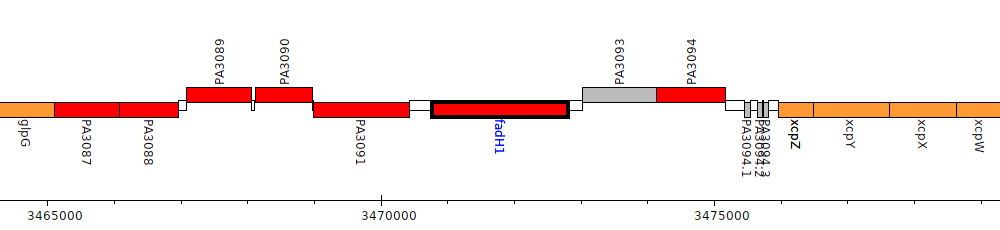
Pseudomonas aeruginosa PAO1, PA3092 (fadH1)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0044255 | cellular lipid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0010181 | FMN binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00724
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016491 | oxidoreductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07992
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.40.50.720 | - | - | - | 378 | 672 | 2.3E-118 |
| CDD | cd02930 | DCR_FMN | - | - | 10 | 362 | 0.0 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 625 | 641 | 1.4E-15 |
| PANTHER | PTHR42917 | 2,4-DIENOYL-COA REDUCTASE | - | - | 7 | 679 | 0.0 |
| PRINTS | PR00411 | Pyridine nucleotide disulphide reductase class-I signature | - | - | 380 | 402 | 2.6E-6 |
| SUPERFAMILY | SSF51971 | Nucleotide-binding domain | - | - | 337 | 526 | 5.12E-43 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 381 | 400 | 1.4E-15 |
| Gene3D | G3DSA:3.20.20.70 | Aldolase class I | IPR013785 | Aldolase-type TIM barrel | 6 | 373 | 4.7E-128 |
| FunFam | G3DSA:3.50.50.60:FF:000113 | NADPH-dependent 2,4-dienoyl-CoA reductase | - | - | 475 | 632 | 4.9E-72 |
| Pfam | PF00724 | NADH:flavin oxidoreductase / NADH oxidase family | IPR001155 | NADH:flavin oxidoreductase/NADH oxidase, N-terminal | 10 | 332 | 6.5E-91 |
| PRINTS | PR00411 | Pyridine nucleotide disulphide reductase class-I signature | - | - | 626 | 640 | 2.6E-6 |
| SUPERFAMILY | SSF51905 | FAD/NAD(P)-binding domain | IPR036188 | FAD/NAD(P)-binding domain superfamily | 466 | 657 | 3.93E-20 |
| SUPERFAMILY | SSF51395 | FMN-linked oxidoreductases | - | - | 4 | 366 | 9.3E-114 |
| Gene3D | G3DSA:3.50.50.60 | - | IPR036188 | FAD/NAD(P)-binding domain superfamily | 475 | 632 | 2.3E-118 |
| FunFam | G3DSA:3.20.20.70:FF:000082 | NADPH-dependent 2,4-dienoyl-CoA reductase | - | - | 6 | 373 | 0.0 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 503 | 521 | 1.4E-15 |
| PRINTS | PR00368 | FAD-dependent pyridine nucleotide reductase signature | - | - | 463 | 481 | 1.4E-15 |
| Pfam | PF07992 | Pyridine nucleotide-disulphide oxidoreductase | IPR023753 | FAD/NAD(P)-binding domain | 379 | 642 | 8.1E-22 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.