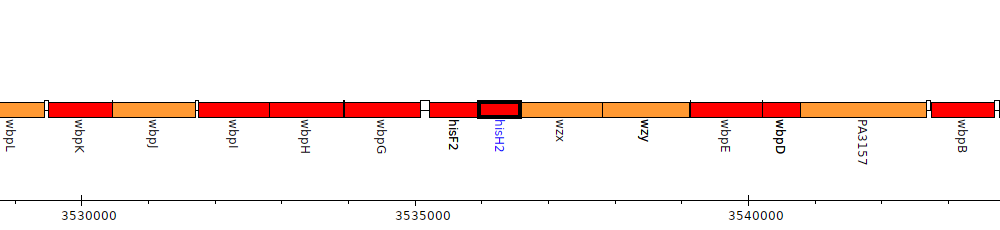
Pseudomonas aeruginosa PAO1, PA3152 (hisH2)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009243 | O antigen biosynthetic process | Inferred from Experiment | ECO:0000269 experimental evidence used in manual assertion |
18621892 | Reviewed by curator |
| Biological Process | GO:0006547 | histidine metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006520 | cellular amino acid metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0000105 | histidine biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000495
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016763 | transferase activity, transferring pentosyl groups |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF000495
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Histidine metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCyc | PWY-7221 | guanosine ribonucleotides de novo biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00340 | Histidine metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | glutaminyl-tRNA<sup>gln</sup> biosynthesis via transamidation | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01230 | Biosynthesis of amino acids | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Hamap | MF_00278 | Imidazole glycerol phosphate synthase subunit HisH [hisH]. | IPR010139 | Imidazole glycerol phosphate synthase, subunit H | 1 | 201 | 34.962563 |
| Pfam | PF00117 | Glutamine amidotransferase class-I | IPR017926 | Glutamine amidotransferase | 3 | 198 | 7.2E-19 |
| SUPERFAMILY | SSF52317 | Class I glutamine amidotransferase-like | IPR029062 | Class I glutamine amidotransferase-like | 2 | 201 | 3.15E-52 |
| NCBIfam | TIGR01855 | JCVI: imidazole glycerol phosphate synthase subunit HisH | IPR010139 | Imidazole glycerol phosphate synthase, subunit H | 2 | 200 | 8.9E-81 |
| Gene3D | G3DSA:3.40.50.880 | - | IPR029062 | Class I glutamine amidotransferase-like | 1 | 202 | 1.4E-74 |
| PANTHER | PTHR42701 | IMIDAZOLE GLYCEROL PHOSPHATE SYNTHASE SUBUNIT HISH | IPR010139 | Imidazole glycerol phosphate synthase, subunit H | 2 | 200 | 3.5E-68 |
| CDD | cd01748 | GATase1_IGP_Synthase | IPR010139 | Imidazole glycerol phosphate synthase, subunit H | 2 | 199 | 4.19347E-112 |
| PIRSF | PIRSF000495 | Amidotransf_HisH | IPR010139 | Imidazole glycerol phosphate synthase, subunit H | 1 | 202 | 4.5E-75 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.