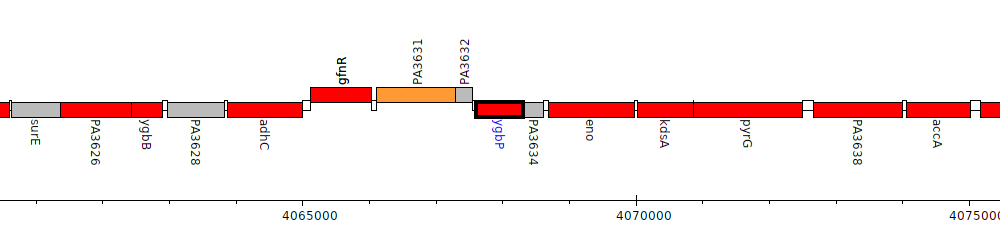
Pseudomonas aeruginosa PAO1, PA3633 (ygbP)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0051188 | obsolete cofactor biosynthetic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0070567 | cytidylyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd02516
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0050518 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00108
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0008299 | isoprenoid biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:MF_00108
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00900 | Terpenoid backbone biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF53448 | Nucleotide-diphospho-sugar transferases | IPR029044 | Nucleotide-diphospho-sugar transferases | 10 | 230 | 5.18E-55 |
| Gene3D | G3DSA:3.90.550.10 | Spore Coat Polysaccharide Biosynthesis Protein SpsA; Chain A | IPR029044 | Nucleotide-diphospho-sugar transferases | 5 | 233 | 5.5E-69 |
| PANTHER | PTHR32125 | 2-C-METHYL-D-ERYTHRITOL 4-PHOSPHATE CYTIDYLYLTRANSFERASE, CHLOROPLASTIC | - | - | 9 | 231 | 8.1E-55 |
| CDD | cd02516 | CDP-ME_synthetase | IPR034683 | Cytidylyltransferase IspD/TarI | 9 | 228 | 4.17242E-82 |
| Hamap | MF_00108 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [ispD]. | IPR001228 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 8 | 232 | 41.353806 |
| NCBIfam | TIGR00453 | JCVI: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | IPR001228 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | 10 | 230 | 1.7E-75 |
| FunFam | G3DSA:3.90.550.10:FF:000003 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | - | - | 7 | 232 | 3.2E-69 |
| Pfam | PF01128 | 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase | IPR034683 | Cytidylyltransferase IspD/TarI | 11 | 227 | 2.6E-64 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.