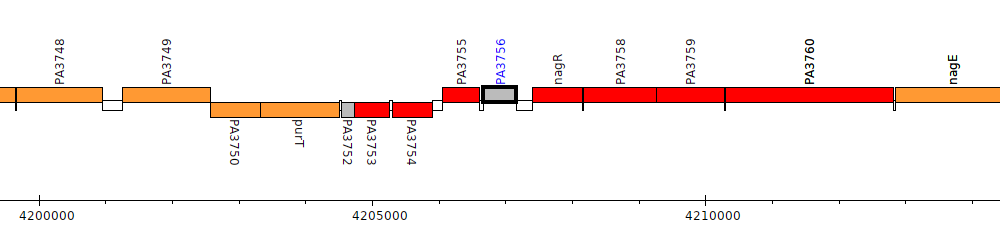
Pseudomonas aeruginosa PAO1, PA3756
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0071972 | peptidoglycan L,D-transpeptidase activity |
Inferred from Sequence or Structural Similarity
Term mapped from: RefSeq:NP_414759.1
|
ECO:0000250 sequence similarity evidence used in manual assertion |
11555281 | Reviewed by curator |
| Molecular Function | GO:0008234 | cysteine-type peptidase activity |
Inferred from Sequence or Structural Similarity
Term mapped from: RefSeq:NP_414759.1
|
ECO:0000250 sequence similarity evidence used in manual assertion |
11555281 | Reviewed by curator |
| Molecular Function | GO:0016740 | transferase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF03734
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF141523 | L,D-transpeptidase catalytic domain-like | IPR038063 | L,D-transpeptidase catalytic domain-like | 30 | 165 | 4.19E-24 |
| CDD | cd16913 | YkuD_like | IPR005490 | L,D-transpeptidase catalytic domain | 31 | 165 | 2.17589E-20 |
| PANTHER | PTHR36699 | LD-TRANSPEPTIDASE | - | - | 18 | 162 | 1.8E-45 |
| Gene3D | G3DSA:2.40.440.10 | - | IPR038063 | L,D-transpeptidase catalytic domain-like | 29 | 164 | 4.0E-19 |
| Pfam | PF03734 | L,D-transpeptidase catalytic domain | IPR005490 | L,D-transpeptidase catalytic domain | 31 | 164 | 1.5E-18 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.