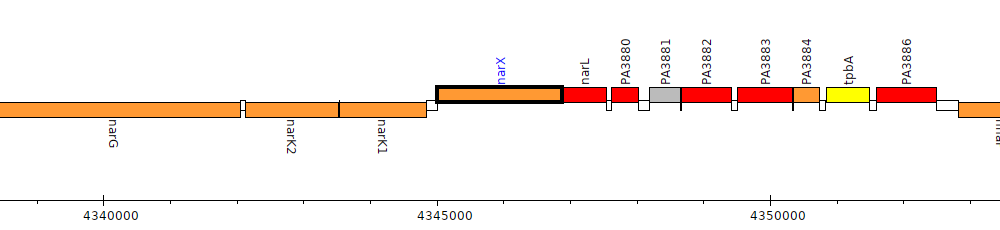
Pseudomonas aeruginosa PAO1, PA3878 (narX)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0008479 | queuine tRNA-ribosyltransferase activity | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006807 | nitrogen compound metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0006091 | generation of precursor metabolites and energy | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0000160 | phosphorelay signal transduction system | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0000160 | phosphorelay signal transduction system |
Inferred from Sequence Model
Term mapped from: InterPro:PF07730
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Cellular Component | GO:0016020 | membrane |
Inferred from Sequence Model
Term mapped from: InterPro:PF00672
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0000155 | phosphorelay sensor kinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07730
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0007165 | signal transduction |
Inferred from Sequence Model
Term mapped from: InterPro:PF00672
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0046983 | protein dimerization activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF07730
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004673 | protein histidine kinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF003167
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Nitrogen metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCAP | Two-component System |
ECO:0000037
not_recorded |
|||
| KEGG | pae02020 | Two-component system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF07730 | Histidine kinase | IPR011712 | Signal transduction histidine kinase, subgroup 3, dimerisation and phosphoacceptor domain | 411 | 476 | 5.4E-17 |
| PANTHER | PTHR24421 | NITRATE/NITRITE SENSOR PROTEIN NARX-RELATED | - | - | 27 | 613 | 5.8E-68 |
| CDD | cd06225 | HAMP | - | - | 195 | 239 | 4.14883E-13 |
| SMART | SM00387 | HKATPase_4 | IPR003594 | Histidine kinase/HSP90-like ATPase | 517 | 611 | 1.8E-9 |
| CDD | cd16917 | HATPase_UhpB-NarQ-NarX-like | - | - | 522 | 603 | 4.04567E-27 |
| Gene3D | G3DSA:1.20.5.1930 | - | - | - | 391 | 473 | 1.4E-14 |
| SUPERFAMILY | SSF158472 | HAMP domain-like | - | - | 193 | 242 | 1.06E-9 |
| Gene3D | G3DSA:3.30.565.10 | - | IPR036890 | Histidine kinase/HSP90-like ATPase superfamily | 477 | 610 | 1.5E-26 |
| Gene3D | G3DSA:1.20.120.960 | Histidine kinase NarX, sensor domain | IPR042295 | NarX-like, N-terminal domain superfamily | 63 | 160 | 5.3E-12 |
| Gene3D | G3DSA:6.10.340.10 | - | - | - | 161 | 251 | 1.7E-14 |
| Coils | Coil | Coil | - | - | 447 | 474 | - |
| Pfam | PF00672 | HAMP domain | IPR003660 | HAMP domain | 192 | 240 | 1.7E-13 |
| PIRSF | PIRSF003167 | HK_NarX/NarQ | IPR016380 | Signal transduction histidine kinase, nitrate/nitrite-sensing | 17 | 622 | 0.0 |
| SUPERFAMILY | SSF55874 | ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase | IPR036890 | Histidine kinase/HSP90-like ATPase superfamily | 508 | 608 | 7.2E-15 |
| SMART | SM00304 | HAMP_11 | IPR003660 | HAMP domain | 192 | 244 | 6.2E-14 |
| Pfam | PF13675 | Type IV pili methyl-accepting chemotaxis transducer N-term | IPR029095 | NarX-like, N-terminal | 62 | 101 | 5.0E-7 |
| Pfam | PF02518 | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase | IPR003594 | Histidine kinase/HSP90-like ATPase | 518 | 608 | 3.6E-8 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.