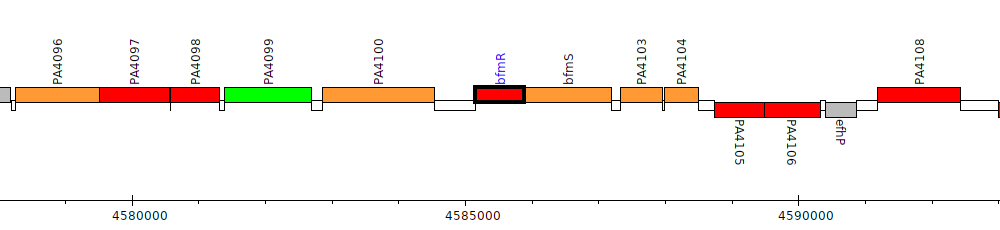
Pseudomonas aeruginosa PAO1, PA4101 (bfmR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Inferred from Direct Assay | ECO:0000096 electrophoretic mobility shift assay evidence |
25166864 | Reviewed by curator |
| Molecular Function | GO:0000156 | phosphorelay response regulator activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
19936057 | Reviewed by curator |
| Biological Process | GO:1900233 | positive regulation of single-species biofilm formation on inanimate substrate | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19936057 | Reviewed by curator |
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:cd00383
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0000160 | phosphorelay signal transduction system |
Inferred from Sequence Model
Term mapped from: InterPro:PF00072
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003677 | DNA binding |
Inferred from Sequence Model
Term mapped from: InterPro:cd00383
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Two-component System |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| SUPERFAMILY | SSF52172 | CheY-like | IPR011006 | CheY-like superfamily | 6 | 133 | 4.41E-40 |
| SMART | SM00862 | Trans_reg_C_3 | IPR001867 | OmpR/PhoB-type DNA-binding domain | 159 | 235 | 2.7E-21 |
| Pfam | PF00486 | Transcriptional regulatory protein, C terminal | IPR001867 | OmpR/PhoB-type DNA-binding domain | 160 | 235 | 2.9E-21 |
| Pfam | PF00072 | Response regulator receiver domain | IPR001789 | Signal transduction response regulator, receiver domain | 7 | 118 | 3.1E-30 |
| SMART | SM00448 | REC_2 | IPR001789 | Signal transduction response regulator, receiver domain | 5 | 117 | 4.2E-41 |
| Gene3D | G3DSA:3.40.50.2300 | - | - | - | 6 | 90 | 2.2E-20 |
| CDD | cd00383 | trans_reg_C | IPR001867 | OmpR/PhoB-type DNA-binding domain | 145 | 235 | 1.06395E-24 |
| FunFam | G3DSA:3.40.50.2300:FF:000001 | DNA-binding response regulator PhoB | - | - | 4 | 85 | 9.0E-25 |
| Gene3D | G3DSA:1.10.10.10 | - | IPR036388 | Winged helix-like DNA-binding domain superfamily | 130 | 237 | 3.1E-28 |
| SUPERFAMILY | SSF46894 | C-terminal effector domain of the bipartite response regulators | IPR016032 | Signal transduction response regulator, C-terminal effector | 135 | 236 | 7.94E-24 |
| Gene3D | G3DSA:6.10.250.690 | - | - | - | 91 | 123 | 2.5E-19 |
| FunFam | G3DSA:1.10.10.10:FF:000099 | Two-component system response regulator TorR | - | - | 129 | 237 | 2.1E-37 |
| PANTHER | PTHR48111 | REGULATOR OF RPOS | IPR039420 | Transcriptional regulatory protein WalR-like | 7 | 236 | 5.9E-50 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.