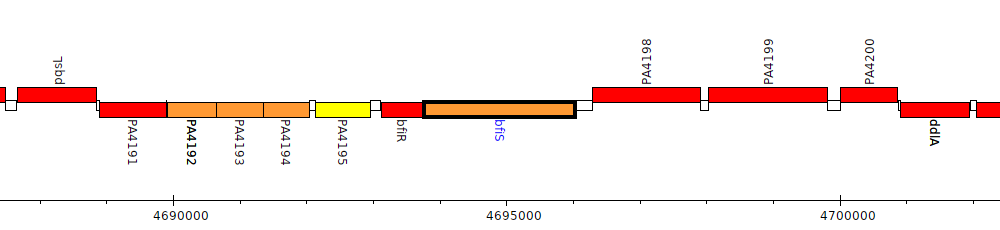
Pseudomonas aeruginosa PAO1, PA4197 (bfiS)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:1900233 | positive regulation of single-species biofilm formation on inanimate substrate | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
19936057 | Reviewed by curator |
| Molecular Function | GO:0000155 | phosphorelay sensor kinase activity | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
19936057 | Reviewed by curator |
| Biological Process | GO:0007165 | signal transduction |
Inferred from Sequence Model
Term mapped from: InterPro:PF00512
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0000155 | phosphorelay sensor kinase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF00512
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016772 | transferase activity, transferring phosphorus-containing groups |
Inferred from Sequence Model
Term mapped from: InterPro:PR00344
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0016310 | phosphorylation |
Inferred from Sequence Model
Term mapped from: InterPro:PR00344
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005515 | protein binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF08447
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| PseudoCAP | Two-component System |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PRINTS | PR00344 | Bacterial sensor protein C-terminal signature | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | 713 | 731 | 1.1E-9 |
| SMART | SM00091 | pas_2 | IPR000014 | PAS domain | 268 | 334 | 2.8E-9 |
| NCBIfam | TIGR00229 | JCVI: PAS domain S-box protein | IPR000014 | PAS domain | 268 | 389 | 1.1E-21 |
| SMART | SM00091 | pas_2 | IPR000014 | PAS domain | 158 | 220 | 66.0 |
| PRINTS | PR00344 | Bacterial sensor protein C-terminal signature | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | 684 | 698 | 1.1E-9 |
| SUPERFAMILY | SSF55785 | PYP-like sensor domain (PAS domain) | IPR035965 | PAS domain superfamily | 273 | 386 | 9.99E-27 |
| Gene3D | G3DSA:3.30.565.10 | - | IPR036890 | Histidine kinase/HSP90-like ATPase superfamily | 599 | 755 | 3.3E-33 |
| SMART | SM00387 | HKATPase_4 | IPR003594 | Histidine kinase/HSP90-like ATPase | 641 | 753 | 3.6E-24 |
| SUPERFAMILY | SSF55874 | ATPase domain of HSP90 chaperone/DNA topoisomerase II/histidine kinase | IPR036890 | Histidine kinase/HSP90-like ATPase superfamily | 590 | 751 | 7.07E-33 |
| SUPERFAMILY | SSF47384 | Homodimeric domain of signal transducing histidine kinase | IPR036097 | Signal transduction histidine kinase, dimerisation/phosphoacceptor domain superfamily | 522 | 597 | 4.71E-7 |
| Pfam | PF02518 | Histidine kinase-, DNA gyrase B-, and HSP90-like ATPase | IPR003594 | Histidine kinase/HSP90-like ATPase | 642 | 751 | 7.3E-15 |
| SMART | SM00086 | pac_2 | IPR001610 | PAC motif | 462 | 504 | 21.0 |
| SMART | SM00086 | pac_2 | IPR001610 | PAC motif | 340 | 383 | 4.8E-7 |
| FunFam | G3DSA:3.30.565.10:FF:000459 | Globin-coupled histidine kinase | - | - | 599 | 755 | 7.8E-92 |
| Gene3D | G3DSA:1.10.287.130 | - | - | - | 531 | 597 | 5.4E-19 |
| CDD | cd00130 | PAS | IPR000014 | PAS domain | 277 | 380 | 7.30277E-16 |
| Coils | Coil | Coil | - | - | 499 | 519 | - |
| PRINTS | PR00344 | Bacterial sensor protein C-terminal signature | IPR004358 | Signal transduction histidine kinase-related protein, C-terminal | 702 | 712 | 1.1E-9 |
| CDD | cd00082 | HisKA | IPR003661 | Signal transduction histidine kinase, dimerisation/phosphoacceptor domain | 524 | 595 | 2.09598E-5 |
| SUPERFAMILY | SSF55785 | PYP-like sensor domain (PAS domain) | IPR035965 | PAS domain superfamily | 382 | 502 | 1.93E-10 |
| SMART | SM00091 | pas_2 | IPR000014 | PAS domain | 391 | 457 | 2.6 |
| SMART | SM00388 | HisKA_10 | IPR003661 | Signal transduction histidine kinase, dimerisation/phosphoacceptor domain | 526 | 599 | 5.6E-5 |
| Pfam | PF00512 | His Kinase A (phospho-acceptor) domain | IPR003661 | Signal transduction histidine kinase, dimerisation/phosphoacceptor domain | 527 | 597 | 3.1E-8 |
| Gene3D | G3DSA:3.30.450.20 | PAS domain | - | - | 401 | 518 | 1.1E-9 |
| PANTHER | PTHR43065 | SENSOR HISTIDINE KINASE | - | - | 317 | 755 | 8.9E-55 |
| CDD | cd00130 | PAS | IPR000014 | PAS domain | 402 | 501 | 8.3341E-4 |
| Pfam | PF08447 | PAS fold | IPR013655 | PAS fold-3 | 291 | 376 | 2.4E-15 |
| Gene3D | G3DSA:3.30.450.20 | PAS domain | - | - | 267 | 400 | 2.3E-30 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.