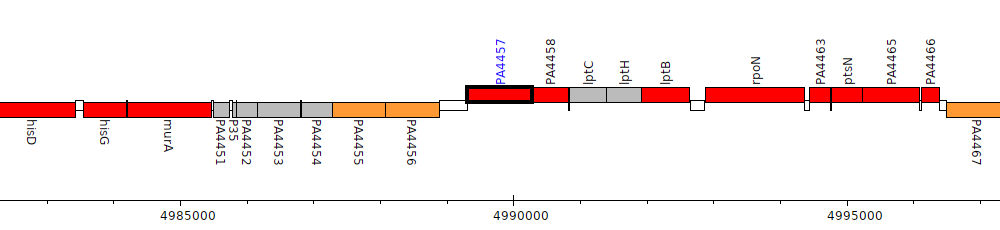
Pseudomonas aeruginosa PAO1, PA4457
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0097367 | carbohydrate derivative binding |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53697
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0005975 | carbohydrate metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF004692
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0016853 | isomerase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF004692
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:1901135 | carbohydrate derivative metabolic process |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53697
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae00540 | Lipopolysaccharide biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR00393 | JCVI: KpsF/GutQ family sugar-phosphate isomerase | IPR004800 | Phosphosugar isomerase, KdsD/KpsF-type | 47 | 316 | 5.6E-113 |
| SMART | SM00116 | cbs_1 | IPR000644 | CBS domain | 277 | 325 | 0.02 |
| SUPERFAMILY | SSF53697 | SIS domain | IPR046348 | SIS domain superfamily | 11 | 324 | 1.69E-69 |
| PIRSF | PIRSF004692 | KdsD_KpsF | IPR004800 | Phosphosugar isomerase, KdsD/KpsF-type | 1 | 326 | 0.0 |
| Gene3D | G3DSA:3.40.50.10490 | - | - | - | 5 | 198 | 6.6E-67 |
| FunFam | G3DSA:3.40.50.10490:FF:000011 | Arabinose 5-phosphate isomerase | - | - | 7 | 197 | 5.8E-87 |
| Pfam | PF00571 | CBS domain | IPR000644 | CBS domain | 219 | 260 | 1.1E-4 |
| SMART | SM00116 | cbs_1 | IPR000644 | CBS domain | 212 | 260 | 3.0 |
| CDD | cd04604 | CBS_pair_SIS_assoc | - | - | 199 | 322 | 2.33601E-64 |
| Gene3D | G3DSA:3.10.580.10 | - | IPR046342 | CBS domain superfamily | 199 | 323 | 1.6E-46 |
| Pfam | PF01380 | SIS domain | IPR001347 | SIS domain | 43 | 174 | 5.0E-32 |
| PANTHER | PTHR42745 | - | - | - | 1 | 326 | 0.0 |
| Pfam | PF00571 | CBS domain | IPR000644 | CBS domain | 279 | 323 | 6.5E-7 |
| FunFam | G3DSA:3.10.580.10:FF:000007 | Arabinose 5-phosphate isomerase | - | - | 199 | 323 | 1.8E-48 |
| CDD | cd05014 | SIS_Kpsf | IPR035474 | KpsF-like, SIS domain | 47 | 174 | 5.41839E-75 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.