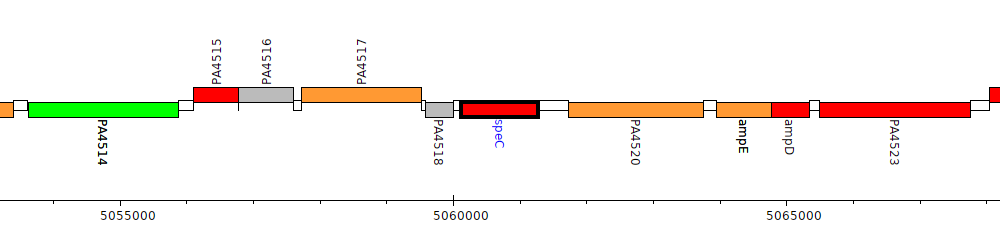
Pseudomonas aeruginosa PAO1, PA4519 (speC)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0033387 | putrescine biosynthetic process from ornithine | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
12634339 | Reviewed by curator |
| Molecular Function | GO:0004586 | ornithine decarboxylase activity | Inferred from Mutant Phenotype | ECO:0000015 mutant phenotype evidence |
12634339 | Reviewed by curator |
| Biological Process | GO:0006596 | polyamine biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PTHR11482
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF02784
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Arginine and proline metabolism |
ECO:0000037
not_recorded |
|||
| PseudoCyc | ORNDEG-PWY | superpathway of ornithine degradation | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pae00330 | Arginine and proline metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | POLYAMSYN-PWY | superpathway of polyamine biosynthesis I | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| PseudoCyc | ORNSPNANA-PWY | ornithine spermine biosynthesis | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCyc | PWY-46 | putrescine biosynthesis III | 19.5 |
ECO:0000250
sequence similarity evidence used in manual assertion |
12867747 |
| KEGG | pae00480 | Glutathione metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.20.20.10 | Alanine racemase | IPR029066 | PLP-binding barrel | 33 | 273 | 1.5E-89 |
| PRINTS | PR01179 | Ornithine/diaminopimelate/arginine (ODA) decarboxylase family signature | IPR000183 | Ornithine/DAP/Arg decarboxylase | 261 | 280 | 3.8E-18 |
| PRINTS | PR01179 | Ornithine/diaminopimelate/arginine (ODA) decarboxylase family signature | IPR000183 | Ornithine/DAP/Arg decarboxylase | 54 | 72 | 3.8E-18 |
| PRINTS | PR01182 | Ornithine decarboxylase signature | IPR002433 | Ornithine decarboxylase | 96 | 120 | 1.1E-18 |
| Gene3D | G3DSA:2.40.37.10 | Lyase, Ornithine Decarboxylase; Chain A, domain 1 | IPR009006 | Alanine racemase/group IV decarboxylase, C-terminal | 21 | 380 | 1.5E-89 |
| PRINTS | PR01182 | Ornithine decarboxylase signature | IPR002433 | Ornithine decarboxylase | 329 | 339 | 1.1E-18 |
| SUPERFAMILY | SSF50621 | Alanine racemase C-terminal domain-like | IPR009006 | Alanine racemase/group IV decarboxylase, C-terminal | 245 | 386 | 1.51E-30 |
| Pfam | PF00278 | Pyridoxal-dependent decarboxylase, C-terminal sheet domain | IPR022643 | Orn/DAP/Arg decarboxylase 2, C-terminal | 272 | 365 | 3.1E-14 |
| PRINTS | PR01179 | Ornithine/diaminopimelate/arginine (ODA) decarboxylase family signature | IPR000183 | Ornithine/DAP/Arg decarboxylase | 177 | 190 | 3.8E-18 |
| PRINTS | PR01182 | Ornithine decarboxylase signature | IPR002433 | Ornithine decarboxylase | 52 | 79 | 1.1E-18 |
| PRINTS | PR01182 | Ornithine decarboxylase signature | IPR002433 | Ornithine decarboxylase | 126 | 148 | 1.1E-18 |
| PRINTS | PR01182 | Ornithine decarboxylase signature | IPR002433 | Ornithine decarboxylase | 26 | 50 | 1.1E-18 |
| Pfam | PF02784 | Pyridoxal-dependent decarboxylase, pyridoxal binding domain | IPR022644 | Orn/DAP/Arg decarboxylase 2, N-terminal | 35 | 271 | 5.4E-55 |
| PANTHER | PTHR11482 | ARGININE/DIAMINOPIMELATE/ORNITHINE DECARBOXYLASE | IPR002433 | Ornithine decarboxylase | 23 | 386 | 1.1E-80 |
| FunFam | G3DSA:2.40.37.10:FF:000004 | Ornithine decarboxylase | - | - | 265 | 380 | 2.4E-59 |
| FunFam | G3DSA:3.20.20.10:FF:000008 | Ornithine decarboxylase | - | - | 34 | 269 | 3.0E-94 |
| PRINTS | PR01179 | Ornithine/diaminopimelate/arginine (ODA) decarboxylase family signature | IPR000183 | Ornithine/DAP/Arg decarboxylase | 74 | 86 | 3.8E-18 |
| CDD | cd00622 | PLPDE_III_ODC | - | - | 25 | 387 | 0.0 |
| SUPERFAMILY | SSF51419 | PLP-binding barrel | IPR029066 | PLP-binding barrel | 25 | 273 | 4.86E-66 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.