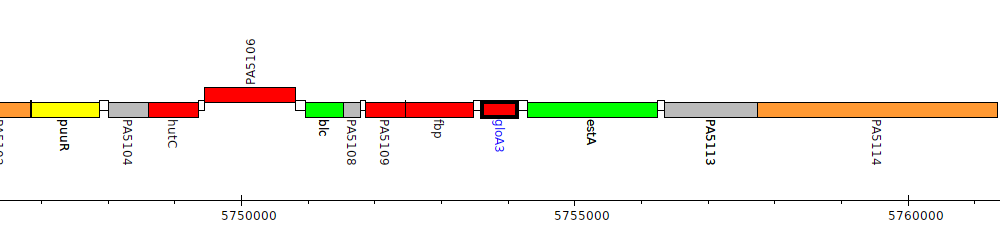
Pseudomonas aeruginosa PAO1, PA5111 (gloA3)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0006090 | pyruvate metabolic process | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
17513180 | Reviewed by curator |
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
17513180 | Reviewed by curator |
| Molecular Function | GO:0008270 | zinc ion binding | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
17513180 | Reviewed by curator |
| Molecular Function | GO:0003961 | O-acetylhomoserine aminocarboxypropyltransferase activity | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Molecular Function | GO:0046872 | metal ion binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00068
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004462 | lactoylglutathione lyase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00068
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pae00620 | Pyruvate metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| PseudoCAP | Pyruvate metabolism |
ECO:0000037
not_recorded |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Gene3D | G3DSA:3.10.180.10 | - | IPR029068 | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | 2 | 169 | 7.7E-63 |
| Pfam | PF00903 | Glyoxalase/Bleomycin resistance protein/Dioxygenase superfamily | IPR004360 | Glyoxalase/fosfomycin resistance/dioxygenase domain | 24 | 164 | 1.5E-23 |
| CDD | cd07233 | GlxI_Zn | - | - | 24 | 165 | 9.05357E-86 |
| NCBIfam | TIGR00068 | JCVI: lactoylglutathione lyase | IPR004361 | Glyoxalase I | 24 | 168 | 2.1E-63 |
| SUPERFAMILY | SSF54593 | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | IPR029068 | Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase | 9 | 167 | 2.82E-37 |
| PANTHER | PTHR10374 | LACTOYLGLUTATHIONE LYASE GLYOXALASE I | - | - | 4 | 169 | 6.3E-75 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.