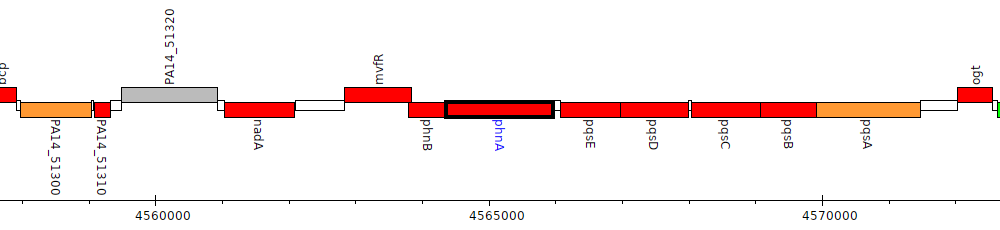
Pseudomonas aeruginosa UCBPP-PA14, PA14_51360 (phnA)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0002047 | phenazine biosynthetic process |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA1001
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
2153661 | |
| Molecular Function | GO:0004049 | anthranilate synthase activity |
Inferred from Sequence or Structural Similarity
Term mapped from: PseudoCAP:PA1001
|
ECO:0000249 sequence similarity evidence used in automatic assertion |
2153661 | |
| Molecular Function | GO:0004049 | anthranilate synthase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00565
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009058 | biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:G3DSA:3.60.120.10
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pau00400 | Phenylalanine, tyrosine and tryptophan biosynthesis | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | acridone alkaloid biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | pau01130 | Biosynthesis of antibiotics | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| MetaCyc | 4-hydroxy-2(1<i>H</i>)-quinolone biosynthesis | InterPro 5.36-75.0 |
ECO:0000259
match to InterPro signature evidence used in automatic assertion |
||
| KEGG | pau02025 | Biofilm formation - Pseudomonas aeruginosa | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau01230 | Biosynthesis of amino acids | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR11236 | AMINOBENZOATE/ANTHRANILATE SYNTHASE | IPR019999 | Anthranilate synthase component I-like | 21 | 522 | 3.8E-107 |
| Pfam | PF04715 | Anthranilate synthase component I, N terminal region | IPR006805 | Anthranilate synthase component I, N-terminal | 48 | 193 | 8.0E-6 |
| PRINTS | PR00095 | Anthranilate synthase component I signature | IPR019999 | Anthranilate synthase component I-like | 447 | 461 | 8.9E-20 |
| SUPERFAMILY | SSF56322 | ADC synthase | IPR005801 | ADC synthase | 36 | 513 | 0.0 |
| Pfam | PF00425 | chorismate binding enzyme | IPR015890 | Chorismate-utilising enzyme, C-terminal | 246 | 505 | 3.5E-79 |
| PIRSF | PIRSF001373 | TrpE | IPR005257 | Anthranilate synthase component I, TrpE-like, bacterial | 6 | 524 | 0.0 |
| PRINTS | PR00095 | Anthranilate synthase component I signature | IPR019999 | Anthranilate synthase component I-like | 367 | 380 | 8.9E-20 |
| PRINTS | PR00095 | Anthranilate synthase component I signature | IPR019999 | Anthranilate synthase component I-like | 353 | 366 | 8.9E-20 |
| Gene3D | G3DSA:3.60.120.10 | Anthranilate synthase | IPR005801 | ADC synthase | 16 | 521 | 0.0 |
| PRINTS | PR00095 | Anthranilate synthase component I signature | IPR019999 | Anthranilate synthase component I-like | 462 | 476 | 8.9E-20 |
| NCBIfam | TIGR00565 | JCVI: anthranilate synthase component I | IPR005257 | Anthranilate synthase component I, TrpE-like, bacterial | 37 | 516 | 0.0 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.