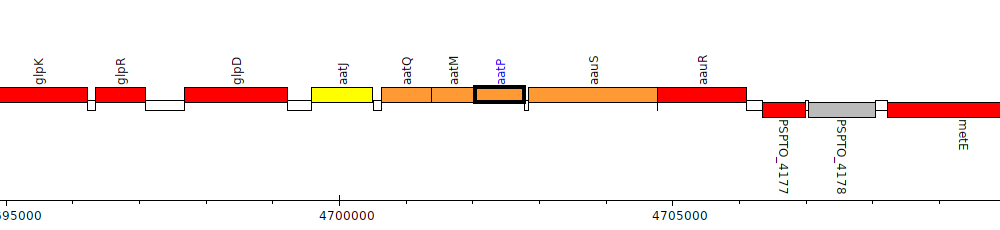
Pseudomonas syringae pv. tomato DC3000, PSPTO_4174 (aatP)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0051938 | L-glutamate import | Inferred from Sequence Orthology | ECO:0000201 sequence orthology evidence |
18310026 | Reviewed by curator |
| Biological Process | GO:0070778 | L-aspartate transmembrane transport |
Inferred from Sequence Orthology
Term mapped from: PseudoCAP:PP_1068
|
ECO:0000201 sequence orthology evidence |
18310026 | Reviewed by curator |
| Cellular Component | GO:0043190 | ATP-binding cassette (ABC) transporter complex | Inferred from Sequence or Structural Similarity | ECO:0000250 sequence similarity evidence used in manual assertion |
18310026 | Reviewed by curator |
| Molecular Function | GO:0016887 | ATPase activity |
Inferred from Sequence Model
Term mapped from: InterPro:SM00382
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00005
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0003333 | amino acid transmembrane transport |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF039085
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0015424 | ATPase-coupled amino acid transmembrane transporter activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF039085
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
|
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pst02020 | Two-component system | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pst02010 | ABC transporters | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.40.50.300:FF:000020 | Amino acid ABC transporter ATP-binding component | - | - | 1 | 242 | 1.0E-120 |
| CDD | cd03262 | ABC_HisP_GlnQ | - | - | 2 | 214 | 0.0 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1 | 226 | 3.67E-76 |
| PIRSF | PIRSF039085 | ABC_ATPase_HisP | IPR030679 | ABC-type amino acid transport system, ATPase component, HisP-type | 1 | 243 | 7.7E-103 |
| PANTHER | PTHR43166 | AMINO ACID IMPORT ATP-BINDING PROTEIN | - | - | 2 | 239 | 4.9E-87 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 1 | 241 | 4.9E-75 |
| SMART | SM00382 | AAA_5 | IPR003593 | AAA+ ATPase domain | 26 | 213 | 6.0E-17 |
| Pfam | PF00005 | ABC transporter | IPR003439 | ABC transporter-like, ATP-binding domain | 17 | 165 | 1.1E-34 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.