Pseudomonas aeruginosa PAO1, PA3961
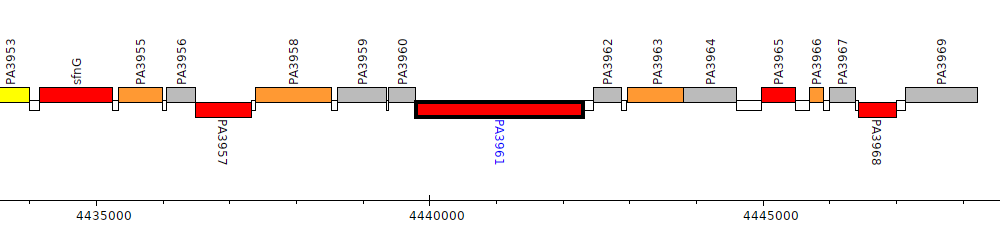
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Molecular Function | GO:0003676 | nucleic acid binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00270
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004386 | helicase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR01970
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005524 | ATP binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00270
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Classifications Manually Assigned by PseudoCAP
|
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| PANTHER | PTHR43519 | ATP-DEPENDENT RNA HELICASE HRPB | - | - | 3 | 837 | 0.0 |
| NCBIfam | TIGR01970 | JCVI: ATP-dependent helicase HrpB | IPR010225 | ATP-dependent helicase HrpB | 4 | 837 | 0.0 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 2 | 182 | 2.6E-54 |
| SMART | SM00490 | helicmild6 | IPR001650 | Helicase, C-terminal | 230 | 334 | 6.5E-19 |
| CDD | cd18791 | SF2_C_RHA | - | - | 184 | 342 | 2.31251E-64 |
| Gene3D | G3DSA:1.20.120.1080 | - | - | - | 378 | 492 | 9.2E-14 |
| CDD | cd17990 | DEXHc_HrpB | - | - | 4 | 179 | 3.61582E-73 |
| Pfam | PF04408 | Helicase associated domain (HA2) | IPR007502 | Helicase-associated domain | 394 | 454 | 4.7E-8 |
| SMART | SM00847 | ha2_5 | IPR007502 | Helicase-associated domain | 393 | 496 | 5.5E-17 |
| SMART | SM00487 | ultradead3 | IPR014001 | Helicase superfamily 1/2, ATP-binding domain | 5 | 189 | 2.1E-17 |
| Pfam | PF00271 | Helicase conserved C-terminal domain | IPR001650 | Helicase, C-terminal | 210 | 333 | 3.1E-14 |
| Gene3D | G3DSA:3.40.50.300 | - | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 183 | 351 | 9.4E-56 |
| Pfam | PF00270 | DEAD/DEAH box helicase | IPR011545 | DEAD/DEAH box helicase domain | 19 | 165 | 5.6E-7 |
| FunFam | G3DSA:1.20.120.1080:FF:000055 | ATP-dependent helicase HrpB | - | - | 377 | 489 | 7.4E-36 |
| SUPERFAMILY | SSF52540 | P-loop containing nucleoside triphosphate hydrolases | IPR027417 | P-loop containing nucleoside triphosphate hydrolase | 30 | 437 | 4.92E-53 |
| PIRSF | PIRSF005496 | ATP_hel_HrpB | IPR010225 | ATP-dependent helicase HrpB | 1 | 838 | 0.0 |
| FunFam | G3DSA:3.40.50.300:FF:002125 | ATP-dependent helicase HrpB | - | - | 1 | 182 | 4.3E-67 |
| Pfam | PF08482 | ATP-dependent helicase C-terminal | IPR013689 | ATP-dependent RNA helicase HrpB, C-terminal | 694 | 825 | 2.4E-52 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.