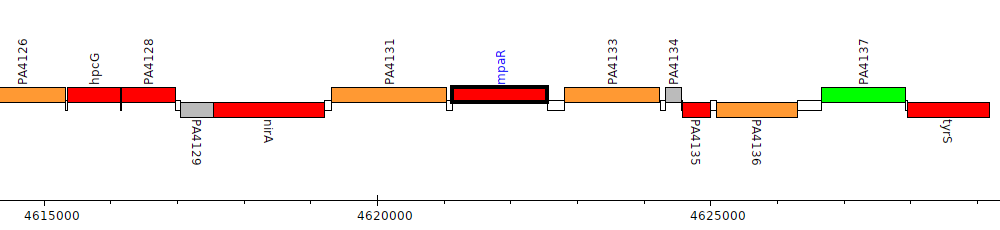
Pseudomonas aeruginosa PAO1, PA4132 (mpaR)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0010628 | positive regulation of gene expression | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
32748556 | Reviewed by curator |
| Biological Process | GO:0062162 | positive regulation of pyocyanine biosynthetic process | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
32748556 | Reviewed by curator |
| Biological Process | GO:1900191 | negative regulation of single-species biofilm formation | Inferred from Mutant Phenotype | ECO:0000315 mutant phenotype evidence used in manual assertion |
32748556 | Reviewed by curator |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | Inferred from Direct Assay | ECO:0001807 |
32748556 | Reviewed by curator |
| Biological Process | GO:0006355 | regulation of transcription, DNA-templated |
Inferred from Sequence Model
Term mapped from: InterPro:cd07377
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0030170 | pyridoxal phosphate binding |
Inferred from Sequence Model
Term mapped from: InterPro:PF00155
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0009058 | biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PF00155
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003700 | DNA-binding transcription factor activity |
Inferred from Sequence Model
Term mapped from: InterPro:cd07377
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| FunFam | G3DSA:3.40.640.10:FF:000023 | Transcriptional regulator, GntR family | - | - | 142 | 361 | 1.3E-79 |
| CDD | cd00609 | AAT_like | - | - | 108 | 464 | 2.62034E-69 |
| Pfam | PF00155 | Aminotransferase class I and II | IPR004839 | Aminotransferase, class I/classII | 167 | 449 | 6.0E-24 |
| SUPERFAMILY | SSF46785 | Winged helix DNA-binding domain | IPR036390 | Winged helix DNA-binding domain superfamily | 2 | 69 | 7.12E-17 |
| Pfam | PF00392 | Bacterial regulatory proteins, gntR family | IPR000524 | Transcription regulator HTH, GntR | 4 | 66 | 1.1E-13 |
| SMART | SM00345 | gntr3 | IPR000524 | Transcription regulator HTH, GntR | 7 | 66 | 9.4E-13 |
| CDD | cd07377 | WHTH_GntR | IPR000524 | Transcription regulator HTH, GntR | 2 | 67 | 1.53871E-18 |
| Gene3D | G3DSA:1.10.10.10 | - | IPR036388 | Winged helix-like DNA-binding domain superfamily | 1 | 67 | 3.9E-17 |
| SUPERFAMILY | SSF53383 | PLP-dependent transferases | IPR015424 | Pyridoxal phosphate-dependent transferase | 98 | 467 | 4.68E-90 |
| PANTHER | PTHR46577 | HTH-TYPE TRANSCRIPTIONAL REGULATORY PROTEIN GABR | - | - | 3 | 466 | 6.3E-86 |
| Gene3D | G3DSA:3.90.1150.10 | Aspartate Aminotransferase, domain 1 | IPR015422 | Pyridoxal phosphate-dependent transferase, small domain | 99 | 464 | 2.0E-93 |
| Gene3D | G3DSA:3.40.640.10 | - | IPR015421 | Pyridoxal phosphate-dependent transferase, major domain | 142 | 361 | 2.0E-93 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.