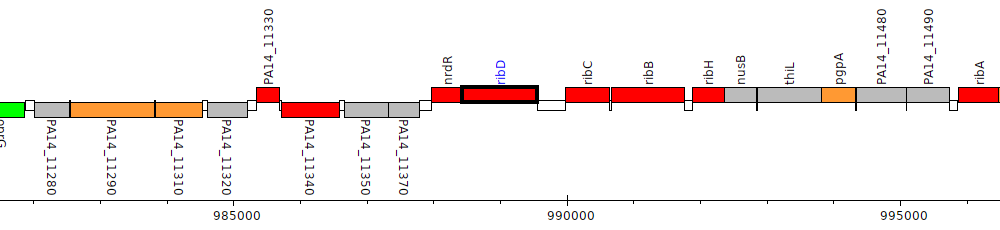
Pseudomonas aeruginosa UCBPP-PA14, PA14_11400 (ribD)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0009231 | riboflavin biosynthetic process |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF006769
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0050661 | NADP binding |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00227
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008703 | 5-amino-6-(5-phosphoribosylamino)uracil reductase activity |
Inferred from Sequence Model
Term mapped from: InterPro:TIGR00227
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0008835 | diaminohydroxyphosphoribosylaminopyrimidine deaminase activity |
Inferred from Sequence Model
Term mapped from: InterPro:PIRSF006769
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0003824 | catalytic activity |
Inferred from Sequence Model
Term mapped from: InterPro:SSF53927
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Pathways
| Database | Xref | Pathway | Version | Evidence | PMID |
|---|---|---|---|---|---|
| KEGG | pau01100 | Metabolic pathways | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau01110 | Biosynthesis of secondary metabolites | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau02024 | Quorum sensing | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
|
| KEGG | pau00740 | Riboflavin metabolism | 81.0+/01-23, Jan 17 |
ECO:0000249
sequence similarity evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| NCBIfam | TIGR00227 | JCVI: riboflavin-specific deaminase C-terminal domain | IPR011549 | Riboflavin-specific deaminase, C-terminal | 148 | 367 | 4.6E-53 |
| Gene3D | G3DSA:3.40.430.10 | Dihydrofolate Reductase, subunit A | IPR024072 | Dihydrofolate reductase-like domain superfamily | 143 | 372 | 2.3E-68 |
| PIRSF | PIRSF006769 | RibD | IPR004794 | Riboflavin biosynthesis protein RibD | 1 | 370 | 4.7E-128 |
| FunFam | G3DSA:3.40.140.10:FF:000025 | Riboflavin biosynthesis protein RibD | - | - | 2 | 142 | 8.5E-68 |
| Pfam | PF00383 | Cytidine and deoxycytidylate deaminase zinc-binding region | IPR002125 | Cytidine and deoxycytidylate deaminase domain | 8 | 101 | 1.3E-19 |
| CDD | cd01284 | Riboflavin_deaminase-reductase | - | - | 9 | 121 | 2.16466E-55 |
| SUPERFAMILY | SSF53597 | Dihydrofolate reductase-like | IPR024072 | Dihydrofolate reductase-like domain superfamily | 147 | 365 | 1.11E-55 |
| Gene3D | G3DSA:3.40.140.10 | Cytidine Deaminase, domain 2 | - | - | 2 | 142 | 2.4E-47 |
| NCBIfam | TIGR00326 | JCVI: bifunctional diaminohydroxyphosphoribosylaminopyrimidine deaminase/5-amino-6-(5-phosphoribosylamino)uracil reductase RibD | IPR004794 | Riboflavin biosynthesis protein RibD | 9 | 364 | 5.0E-116 |
| Pfam | PF01872 | RibD C-terminal domain | IPR002734 | Bacterial bifunctional deaminase-reductase, C-terminal | 149 | 363 | 3.3E-47 |
| SUPERFAMILY | SSF53927 | Cytidine deaminase-like | IPR016193 | Cytidine deaminase-like | 5 | 146 | 7.03E-49 |
| PANTHER | PTHR38011 | DIHYDROFOLATE REDUCTASE FAMILY PROTEIN (AFU_ORTHOLOGUE AFUA_8G06820) | - | - | 142 | 366 | 2.2E-33 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.