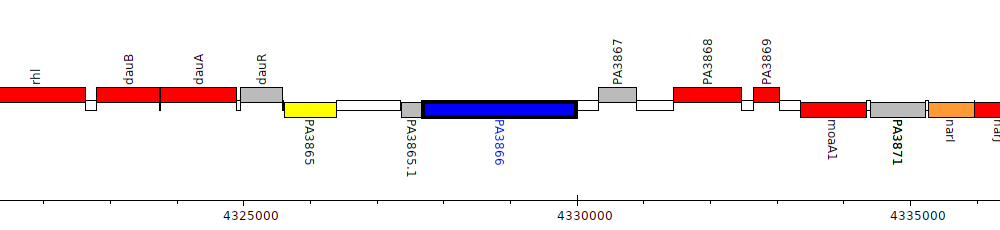
Pseudomonas aeruginosa PAO1, PA3866
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
Gene Ontology
| Ontology | Accession | Term | GO Evidence | Evidence Ontology (ECO) Code | Reference | Comments |
|---|---|---|---|---|---|---|
| Biological Process | GO:0019835 | cytolysis | Inferred from Direct Assay | ECO:0000314 direct assay evidence used in manual assertion |
7721800 | Reviewed by curator |
| Biological Process | GO:0050896 | response to stimulus | Inferred from Sequence or Structural Similarity | ECO:0000044 sequence similarity evidence |
||
| Biological Process | GO:0009617 | response to bacterium |
Inferred from Sequence Model
Term mapped from: InterPro:PR01300
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004540 | ribonuclease activity |
Inferred from Sequence Model
Term mapped from: InterPro:PF12106
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Biological Process | GO:0019835 | cytolysis |
Inferred from Sequence Model
Term mapped from: InterPro:PR01300
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0004519 | endonuclease activity |
Inferred from Sequence Model
Term mapped from: InterPro:PR01300
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
||
| Molecular Function | GO:0005102 | signaling receptor binding |
Inferred from Sequence Model
Term mapped from: InterPro:PR01300
|
ECO:0000259 match to InterPro signature evidence used in automatic assertion |
Functional Predictions from Interpro
| Analysis | Accession | Description | Interpro Accession | Interpro Description | Amino Acid Start | Amino Acid Stop | E-value |
|---|---|---|---|---|---|---|---|
| Pfam | PF06958 | S-type Pyocin | IPR016128 | Pyosin/cloacin translocation domain | 519 | 663 | 3.1E-36 |
| PRINTS | PR01300 | Pyocin S killer protein signature | IPR003060 | Pyocin S killer protein | 420 | 441 | 4.1E-10 |
| Pfam | PF12106 | Colicin E5 ribonuclease domain | IPR021964 | Colicin E5 ribonuclease domain | 669 | 757 | 6.4E-37 |
| Coils | Coil | Coil | - | - | 329 | 349 | - |
| MobiDBLite | mobidb-lite | consensus disorder prediction | - | - | 697 | 721 | - |
| Gene3D | G3DSA:3.30.2310.30 | - | IPR038234 | Colicin E5 ribonuclease domain superfamily | 663 | 764 | 1.4E-31 |
| Coils | Coil | Coil | - | - | 381 | 405 | - |
| SUPERFAMILY | SSF69369 | Cloacin translocation domain | IPR036302 | Pyosin/cloacin translocation domain superfamily | 457 | 692 | 6.02E-49 |
| Coils | Coil | Coil | - | - | 64 | 87 | - |
| Coils | Coil | Coil | - | - | 182 | 216 | - |
| SUPERFAMILY | SSF102824 | Colicin D/E5 nuclease domain | IPR038233 | Colicin D/E5 nuclease domain superfamily | 671 | 763 | 2.56E-35 |
| PRINTS | PR01300 | Pyocin S killer protein signature | IPR003060 | Pyocin S killer protein | 466 | 484 | 4.1E-10 |
| PRINTS | PR01300 | Pyocin S killer protein signature | IPR003060 | Pyocin S killer protein | 501 | 524 | 4.1E-10 |
Search for additional functional domains at the NCBI CDD database website. Go to this protein's amino acid sequence and follow the link.