Pseudomonas aeruginosa PAO1, PA2014 (liuB)
Cytoplasmic
Cytoplasmic Membrane
Periplasmic
Outer Membrane
Extracellular
Unknown
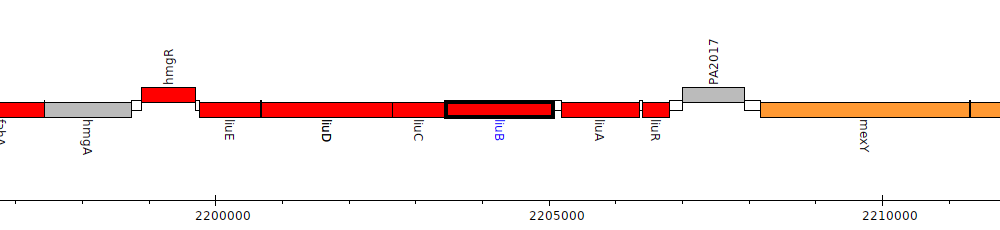
Gene Feature Overview
| Strain |
Pseudomonas aeruginosa PAO1 (Stover et al., 2000)
GCF_000006765.1|latest |
| Locus Tag |
PA2014
|
| Name |
liuB
Synonym: gnyB |
| Replicon | chromosome |
| Genomic location | 2203460 - 2205067 (- strand) |
| Transposon Mutants in orthologs | 2 transposon mutants in orthologs |
Cross-References
| RefSeq | NP_250704.1 |
| GI | 15597210 |
| Affymetrix | PA2014_at |
| Entrez | 878244 |
| GenBank | AAG05402.1 |
| INSDC | AAG05402.1 |
| NCBI Locus Tag | PA2014 |
| protein_id(GenBank) | gb|AAG05402.1|AE004627_10|gnl|PseudoCAP|PA2014 |
| TIGR | NTL03PA02014 |
| UniParc | UPI00000C555A |
| UniProtKB Acc | Q9I297 |
| UniProtKB ID | Q9I297_PSEAE |
| UniRef100 | UniRef100_W0YNW0 |
| UniRef50 | UniRef50_Q9LDD8 |
| UniRef90 | UniRef90_A0A024HIF6 |
Product
| Feature Type | CDS |
| Coding Frame | 1 |
| Product Name |
methylcrotonyl-CoA carboxylase, beta-subunit
|
| Synonyms |
MccB beta subunit of geranoyl-CoA carboxylase, GnyB |
| Evidence for Translation |
Identified using nanoflow high-pressure liquid chromatography (HPLC) in conjunction with microelectrospray ionization on LTQ XL mass spectrometer (PMID:24291602).
|
| Charge (pH 7) | -2.68 |
| Kyte-Doolittle Hydrophobicity Value | 0.021 |
| Molecular Weight (kDa) | 57.4 |
| Isoelectric Point (pI) | 6.67 |
Subcellular localization
| Individual Mappings | |
| Additional evidence for subcellular localization |
PDB 3D Structures
| Accession | Header | Accession Date | Compound | Source | Resolution | Method | Percent Identity |
| 3U9T | LIGASE | 10/19/11 | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, free enzyme | Pseudomonas aeruginosa | 2.9 | X-RAY DIFFRACTION | 100.0 |
| 3U9R | LIGASE | 10/19/11 | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC), beta subunit | Pseudomonas aeruginosa | 1.5 | X-RAY DIFFRACTION | 100.0 |
| 3U9S | LIGASE | 10/19/11 | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | Pseudomonas aeruginosa | 3.5 | X-RAY DIFFRACTION | 100.0 |
Pathogen Association Analysis
| Results |
Common
Found in both pathogen and nonpathogenic strains
Hits to this gene were found in 398 genera
|
Orthologs/Comparative Genomics
| Pseudomonas Ortholog Database | View orthologs at Pseudomonas Ortholog Database |
| Pseudomonas Ortholog Group |
POG003425 (534 members) |
| Putative Inparalogs | None Found |
Interactions
| STRING database | Search for predicted protein-protein interactions using:
Search term: PA2014
Search term: liuB
Search term: methylcrotonyl-CoA carboxylase, beta-subunit
|
Human Homologs
|
Ensembl
110, assembly
GRCh38.p14
|
methylcrotonyl-CoA carboxylase subunit 2 [Source:HGNC Symbol;Acc:HGNC:6937]
E-value:
9.3e-206
Percent Identity:
65.3
|
References
|
The gnyRDBHAL cluster is involved in acyclic isoprenoid degradation in Pseudomonas aeruginosa.
Díaz-Pérez AL, Zavala-Hernández AN, Cervantes C, Campos-García J
Appl Environ Microbiol 2004 Sep;70(9):5102-10
PubMed ID: 15345388
|
|
Identification of genes and proteins necessary for catabolism of acyclic terpenes and leucine/isovalerate in Pseudomonas aeruginosa.
Förster-Fromme K, Höschle B, Mack C, Bott M, Armbruster W, Jendrossek D
Appl Environ Microbiol 2006 Jul;72(7):4819-28
PubMed ID: 16820476
|
|
An integrated map of the genome of the tubercle bacillus, Mycobacterium tuberculosis H37Rv, and comparison with Mycobacterium leprae.
Philipp WJ, Poulet S, Eiglmeier K, Pascopella L, Balasubramanian V, Heym B, Bergh S, Bloom BR, Jacobs WR Jr, Cole ST
Proc Natl Acad Sci U S A 1996 Apr 2;93(7):3132-7
PubMed ID: 8610181
|